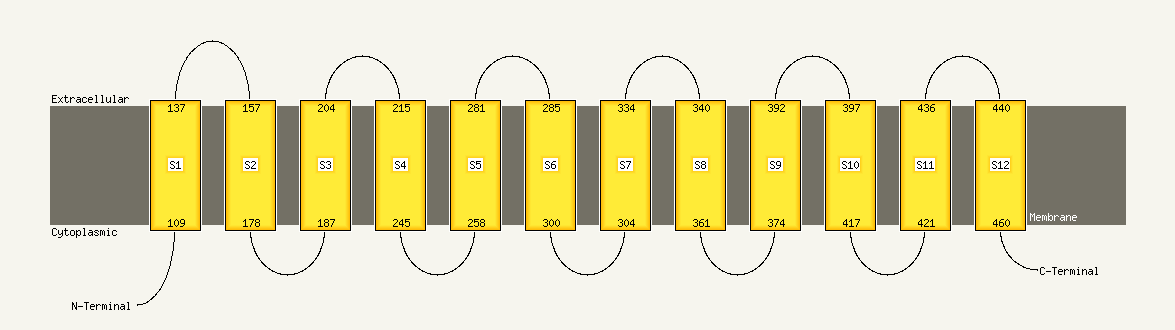
1 c3qnqD_
100.0
12
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
2 c3ipjB_
99.9
27
PDB header: transferaseChain: B: PDB Molecule: pts system, iiabc component;PDBTitle: the crystal structure of one domain of the pts system, iiabc component2 from clostridium difficile
3 c1ibaA_
99.8
22
PDB header: phoshphotransferaseChain: A: PDB Molecule: glucose permease;PDBTitle: glucose permease (domain iib), nmr, 11 structures
4 d3bp8c1
99.8
21
Fold: Homing endonuclease-likeSuperfamily: Glucose permease domain IIBFamily: Glucose permease domain IIB5 c2voyB_
48.2
18
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
6 c3sy6A_
44.2
21
PDB header: cell adhesionChain: A: PDB Molecule: fimbrial protein bf1861;PDBTitle: crystal structure of a fimbrial protein bf1861 [bacteroides fragilis2 nctc 9343] (bf1861) from bacteroides fragilis nctc 9343 at 1.90 a3 resolution
7 c3t2lA_
39.2
16
PDB header: cell adhesionChain: A: PDB Molecule: putative cell adhesion protein;PDBTitle: crystal structure of a putative cell adhesion protein (bf1858) from2 bacteroides fragilis nctc 9343 at 2.33 a resolution
8 c2janD_
29.3
24
PDB header: ligaseChain: D: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: tyrosyl-trna synthetase from mycobacterium tuberculosis in2 unliganded state
9 c2kncB_
25.7
14
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
10 c2kncA_
25.4
6
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
11 d1ffgb_
23.5
10
Fold: Ferredoxin-likeSuperfamily: CheY-binding domain of CheAFamily: CheY-binding domain of CheA12 c1a0oH_
22.6
10
PDB header: chemotaxisChain: H: PDB Molecule: chea;PDBTitle: chey-binding domain of chea in complex with chey
13 c2pmuD_
19.4
15
PDB header: transcription regulationChain: D: PDB Molecule: response regulator phop;PDBTitle: crystal structure of the dna-binding domain of phop
14 c2hqnA_
18.8
12
PDB header: signaling proteinChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structure of a atypical orphan response regulator protein revealed a2 new phosphorylation-independent regulatory mechanism
15 c1m6yA_
17.3
11
PDB header: transferaseChain: A: PDB Molecule: s-adenosyl-methyltransferase mraw;PDBTitle: crystal structure analysis of tm0872, a putative sam-2 dependent methyltransferase, complexed with sah
16 d1pu6a_
17.0
4
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: 3-Methyladenine DNA glycosylase III (MagIII)17 d2bgwa1
16.8
23
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like18 d1vgga_
16.4
22
Fold: Ta1353-likeSuperfamily: Ta1353-likeFamily: Ta1353-like19 d1kgsa1
16.2
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like20 d1vr6a1
15.2
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase21 d1ciya1
not modelled
15.2
26
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain22 d2cyya2
not modelled
15.0
11
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain23 d1gxqa_
not modelled
14.9
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like24 d2gola1
not modelled
14.6
8
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: Immunodeficiency virus matrix proteins25 c3zqsB_
not modelled
14.6
11
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase fancl;PDBTitle: human fancl central domain
26 c1oy8A_
not modelled
14.6
17
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
27 c2wshC_
not modelled
14.5
10
PDB header: hydrolaseChain: C: PDB Molecule: endonuclease ii;PDBTitle: structure of bacteriophage t4 endoii e118a mutant
28 d1ji6a1
not modelled
12.9
26
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain29 c3k8hA_
not modelled
12.5
25
PDB header: membrane proteinChain: A: PDB Molecule: 30klp;PDBTitle: structure of crystal form i of tp0453
30 d2cfua2
not modelled
12.3
21
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Alkylsulfatase-like31 c1vbkA_
not modelled
11.8
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ph1313;PDBTitle: crystal structure of ph1313 from pyrococcus horikoshii ot3
32 c3ufiA_
not modelled
10.9
17
PDB header: cell adhesionChain: A: PDB Molecule: hypothetical protein bacova_04980;PDBTitle: crystal structure of a hypothetical protein2 bacova_04980(zp_02067969.1) from bacteroides ovatus atcc 8483 at 2.183 a resolution
33 c2cfuA_
not modelled
10.8
21
PDB header: hydrolaseChain: A: PDB Molecule: sdsa1;PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
34 d1qu3a1
not modelled
10.7
10
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases35 d2a5la1
not modelled
10.2
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like36 d1wlqc_
not modelled
10.1
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DNA replication factor Cdt137 d1mwza_
not modelled
10.1
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain38 c3ibwA_
not modelled
10.0
8
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
39 c2jnhA_
not modelled
10.0
7
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of the uba domain from cbl-b
40 c2ekmC_
not modelled
9.8
30
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein st1511;PDBTitle: structure of st1219 protein from sulfolobus tokodaii
41 d2r7da2
not modelled
9.8
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RNB domain-like42 c2gl0A_
not modelled
9.6
33
PDB header: transferaseChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: structure of pae2307 in complex with adenosine
43 d1fzda_
not modelled
9.6
22
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like44 d1l6na1
not modelled
9.4
8
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: Immunodeficiency virus matrix proteins45 c2j61B_
not modelled
9.0
26
PDB header: lectinChain: B: PDB Molecule: ficolin-2;PDBTitle: l-ficolin complexed to n-acetylglucosamine (forme c)
46 d1i1ga2
not modelled
8.4
10
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain47 d2prra1
not modelled
8.3
24
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like48 c2jzyA_
not modelled
8.2
19
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein pcor;PDBTitle: solution structure of c-terminal effector domain of2 putative two-component-system response regulator involved3 in copper resistance from klebsiella pneumoniae
49 c2bbjB_
not modelled
8.2
13
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
50 d1tama_
not modelled
8.1
8
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: Immunodeficiency virus matrix proteins51 c2k5eA_
not modelled
8.1
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
52 d1od5a2
not modelled
7.9
22
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein53 d1lwub1
not modelled
7.7
13
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like54 d1hiwa_
not modelled
7.7
4
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: Immunodeficiency virus matrix proteins55 c2xv4S_
not modelled
7.6
15
PDB header: transcriptionChain: S: PDB Molecule: dna-directed rna polymerase iii subunit rpc3;PDBTitle: structure of human rpc62 (partial)
56 c2do6A_
not modelled
7.6
7
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of rsgi ruh-065, a uba domain from human2 cdna
57 d1m1jc1
not modelled
7.3
26
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like58 d1ffya1
not modelled
7.3
12
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases59 d1rrqa1
not modelled
7.3
11
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: Mismatch glycosylase60 c2aj1A_
not modelled
7.3
13
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
61 d1x0pa1
not modelled
7.2
24
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: BLUF domain62 d2a1jb1
not modelled
7.2
27
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like63 d1ngka_
not modelled
7.2
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Truncated hemoglobin64 c2d9sA_
not modelled
7.2
7
PDB header: ligaseChain: A: PDB Molecule: cbl e3 ubiquitin protein ligase;PDBTitle: solution structure of rsgi ruh-049, a uba domain from mouse2 cdna
65 d1j9ia_
not modelled
7.1
8
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Terminase gpNU1 subunit domain66 d1x2ia1
not modelled
7.0
26
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like67 d1orna_
not modelled
7.0
17
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: Endonuclease III68 d1j4na_
not modelled
7.0
14
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like69 d1dlca1
not modelled
7.0
39
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain70 c2kscA_
not modelled
6.9
25
PDB header: unknown functionChain: A: PDB Molecule: cyanoglobin;PDBTitle: solution structure of synechococcus sp. pcc 7002 hemoglobin
71 c1dlcA_
not modelled
6.9
32
PDB header: toxinChain: A: PDB Molecule: delta-endotoxin cryiiia;PDBTitle: crystal structure of insecticidal delta-endotoxin from2 bacillus thuringiensis at 2.5 angstroms resolution
72 d1fida_
not modelled
6.9
30
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like73 c2e1cA_
not modelled
6.9
10
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
74 c1p68A_
not modelled
6.7
29
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed protein s-824;PDBTitle: solution structure of s-824, a de novo designed four helix2 bundle
75 d1fftb2
not modelled
6.6
12
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region76 c2zbcH_
not modelled
6.6
13
PDB header: transcriptionChain: H: PDB Molecule: 83aa long hypothetical transcriptional regulator asnc;PDBTitle: crystal structure of sts042, a stand-alone ram module protein, from2 hyperthermophilic archaeon sulfolobus tokodaii strain7.
77 c2r7fA_
not modelled
6.6
14
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease ii family protein;PDBTitle: crystal structure of ribonuclease ii family protein from deinococcus2 radiodurans, hexagonal crystal form. northeast structural genomics3 target drr63
78 d1m1ha2
not modelled
6.5
13
Fold: Ferredoxin-likeSuperfamily: N-utilization substance G protein NusG, N-terminal domainFamily: N-utilization substance G protein NusG, N-terminal domain79 c3aq8A_
not modelled
6.5
17
PDB header: oxygen bindingChain: A: PDB Molecule: group 1 truncated hemoglobin;PDBTitle: crystal structure of truncated hemoglobin from tetrahymena pyriformis,2 q46e mutant, fe(iii) form
80 c2djwF_
not modelled
6.4
25
PDB header: unknown functionChain: F: PDB Molecule: probable transcriptional regulator, asnc family;PDBTitle: crystal structure of ttha0845 from thermus thermophilus hb8
81 c3pcoD_
not modelled
6.4
15
PDB header: ligaseChain: D: PDB Molecule: phenylalanyl-trna synthetase, beta chain;PDBTitle: crystal structure of e. coli phenylalanine-trna synthetase complexed2 with phenylalanine and amp
82 c2w1oA_
not modelled
6.4
25
PDB header: translationChain: A: PDB Molecule: 60s acidic ribosomal protein p2;PDBTitle: nmr structure of dimerization domain of human ribosomal2 protein p2
83 c2jsxA_
not modelled
6.4
8
PDB header: chaperoneChain: A: PDB Molecule: protein napd;PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
84 d1s69a_
not modelled
6.3
25
Fold: Globin-likeSuperfamily: Globin-likeFamily: Truncated hemoglobin85 d2csba3
not modelled
6.2
17
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain86 d1ilea1
not modelled
6.2
15
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases87 c2hfnJ_
not modelled
6.2
24
PDB header: electron transportChain: J: PDB Molecule: synechocystis photoreceptor (slr1694);PDBTitle: crystal structures of the synechocystis photoreceptor slr1694 reveal2 distinct structural states related to signaling
88 d2vv5a3
not modelled
6.2
10
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane regionSuperfamily: Mechanosensitive channel protein MscS (YggB), transmembrane regionFamily: Mechanosensitive channel protein MscS (YggB), transmembrane region89 c3bvhC_
not modelled
6.1
30
PDB header: blood clottingChain: C: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of recombinant gammad364a fibrinogen fragment d with2 the peptide ligand gly-pro-arg-pro-amide
90 d1jida_
not modelled
6.1
40
Fold: SRP19Superfamily: SRP19Family: SRP1991 d1vpba_
not modelled
6.0
13
Fold: Putative modulator of DNA gyrase, PmbA/TldDSuperfamily: Putative modulator of DNA gyrase, PmbA/TldDFamily: Putative modulator of DNA gyrase, PmbA/TldD92 d1rzsa_
not modelled
6.0
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors93 c1jb7A_
not modelled
6.0
14
PDB header: dna-binding protein/dnaChain: A: PDB Molecule: telomere-binding protein alpha subunit;PDBTitle: dna g-quartets in a 1.86 a resolution structure of an oxytricha nova2 telomeric protein-dna complex
94 c1ph4A_
not modelled
6.0
14
PDB header: dna binding protein/dnaChain: A: PDB Molecule: telomere-binding protein alpha subunit;PDBTitle: crystal structure of the oxytricha nova telomere end-binding protein2 complexed with noncognate ssdna ggggttttggcg
95 d1f7ua3
not modelled
6.0
12
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domainFamily: Arginyl-tRNA synthetase (ArgRS), N-terminal 'additional' domain96 c2jvaA_
not modelled
6.0
23
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase domain protein;PDBTitle: nmr solution structure of peptidyl-trna hydrolase domain protein from2 pseudomonas syringae pv. tomato. northeast structural genomics3 consortium target psr211
97 d2cfxa2
not modelled
6.0
25
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain98 d1re3b1
not modelled
5.9
17
Fold: Fibrinogen C-terminal domain-likeSuperfamily: Fibrinogen C-terminal domain-likeFamily: Fibrinogen C-terminal domain-like99 c2kddB_
not modelled
5.8
31
PDB header: cell cycleChain: B: PDB Molecule: borealin;PDBTitle: solution structure of the conserved c-terminal dimerization2 domain of borealin