1 c2hdiA_
100.0
95
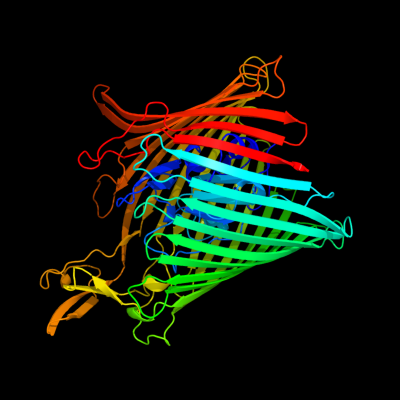
PDB header: protein transport,antimicrobial proteinChain: A: PDB Molecule: colicin i receptor;PDBTitle: crystal structure of the colicin i receptor cir from e.coli in complex2 with receptor binding domain of colicin ia.
2 c2iahA_
100.0
15
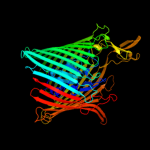
PDB header: membrane proteinChain: A: PDB Molecule: ferripyoverdine receptor;PDBTitle: crystal structure of the ferripyoverdine receptor of the outer2 membrane of pseudomonas aeruginosa bound to ferripyoverdine.
3 c3qlbA_
100.0
15
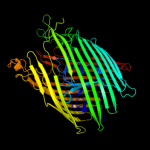
PDB header: metal transportChain: A: PDB Molecule: enantio-pyochelin receptor;PDBTitle: enantiopyochelin outer membrane tonb-dependent transporter from2 pseudomonas fluorescens bound to the ferri-enantiopyochelin
4 c1xkhC_
100.0
14
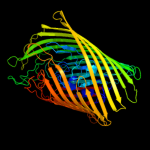
PDB header: membrane proteinChain: C: PDB Molecule: ferripyoverdine receptor;PDBTitle: pyoverdine outer membrane receptor fpva from pseudomonas aeruginosa2 pao1 bound to pyoverdine
5 d1by5a_
100.0
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel6 c1xkwA_
100.0
13
PDB header: membrane proteinChain: A: PDB Molecule: fe(iii)-pyochelin receptor;PDBTitle: pyochelin outer membrane receptor fpta from pseudomonas2 aeruginosa
7 c2grxB_
100.0
14
PDB header: metal transportChain: B: PDB Molecule: ferrichrome-iron receptor;PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
8 c3fhhA_
100.0
19
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane heme receptor shua;PDBTitle: crystal structure of the heme/hemoglobin outer membrane2 transporter shua from shigella dysenteriae
9 d1fepa_
100.0
29
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel10 c3efmA_
100.0
13
PDB header: membrane proteinChain: A: PDB Molecule: ferric alcaligin siderophore receptor;PDBTitle: structure of the alcaligin outer membrane recepteur faua from2 bordetella pertussis
11 d2gufa1
100.0
25
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel12 d1kmoa_
100.0
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel13 c1po3A_
100.0
14
PDB header: membrane proteinChain: A: PDB Molecule: iron(iii) dicitrate transport protein fecaPDBTitle: crystal structure of ferric citrate transporter feca in2 complex with ferric citrate
14 c3cslB_
100.0
16
PDB header: membrane protein/heme binding proteinChain: B: PDB Molecule: hasr protein;PDBTitle: structure of the serratia marcescens hemophore receptor hasr in2 complex with its hemophore hasa and heme
15 d1t16a_
99.1
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein16 c3dwoX_
98.0
8
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
17 c2f1tB_
97.4
11
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
18 c3qraA_
97.0
16
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
19 c3brzA_
96.6
11
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
20 c2k0lA_
96.4
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
21 d1qjpa_
not modelled
95.9
15
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein22 d1g90a_
not modelled
95.9
11
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein23 c3nb3C_
not modelled
95.7
12
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
24 c2iwvD_
not modelled
94.6
14
PDB header: ion channelChain: D: PDB Molecule: outer membrane protein g;PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
25 c2wjqA_
not modelled
94.5
15
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
26 c2x27X_
not modelled
94.4
17
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
27 d1p4ta_
not modelled
92.4
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein28 d1qj8a_
not modelled
91.2
11
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein29 c2jmmA_
not modelled
89.8
12
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
30 d2fgqx1
not modelled
87.8
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin31 d1uynx_
not modelled
80.3
11
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter32 c3bryB_
not modelled
77.2
10
PDB header: transport proteinChain: B: PDB Molecule: tbux;PDBTitle: crystal structure of the ralstonia pickettii toluene2 transporter tbux
33 c2lhfA_
not modelled
71.6
9
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
34 d1osma_
not modelled
68.3
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin35 c3nsgA_
not modelled
54.5
14
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
36 c3kvnA_
not modelled
51.6
15
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
37 d1phoa_
not modelled
50.9
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin38 c3qq2C_
not modelled
45.5
8
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
39 c2y0kA_
not modelled
39.9
14
PDB header: transport proteinChain: A: PDB Molecule: pyroglutatmate porin opdo;PDBTitle: crystal structure of pseudomonas aeruginosa opdo
40 d2zfga1
not modelled
34.3
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin41 c3a2rX_
not modelled
27.0
13
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
42 c3aehB_
not modelled
24.2
15
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
43 c2qomB_
not modelled
22.8
10
PDB header: hydrolaseChain: B: PDB Molecule: serine protease espp;PDBTitle: the crystal structure of the e.coli espp autotransporter beta-domain.
44 c3sljA_
not modelled
22.6
10
PDB header: protein transportChain: A: PDB Molecule: serine protease espp;PDBTitle: pre-cleavage structure of the autotransporter espp - n1023a mutant
45 c2x4mD_
not modelled
22.5
16
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
46 d2a1ja1
not modelled
19.9
50
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like47 c2y0hA_
not modelled
17.4
14
PDB header: transport proteinChain: A: PDB Molecule: probable porin;PDBTitle: crystal structure of pseudomonas aeruginosa opdl
48 c2eg9B_
not modelled
16.3
5
PDB header: hydrolaseChain: B: PDB Molecule: adp-ribosyl cyclase 1;PDBTitle: crystal structure of the truncated extracellular domain of2 mouse cd38
49 c1oy8A_
not modelled
14.4
32
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
50 d3prna_
not modelled
12.1
20
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin51 d2i4sa1
not modelled
11.7
13
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like52 c2kl1A_
not modelled
8.6
16
PDB header: protein bindingChain: A: PDB Molecule: ylbl protein;PDBTitle: solution structure of gtr34c from geobacillus thermodenitrificans.2 northeast structural genomics consortium target gtr34c
53 c3k07A_
not modelled
8.4
27
PDB header: transport proteinChain: A: PDB Molecule: cation efflux system protein cusa;PDBTitle: crystal structure of cusa
54 c3lkxB_
not modelled
8.3
43
PDB header: chaperoneChain: B: PDB Molecule: nascent polypeptide-associated complex subunit alpha;PDBTitle: human nac dimerization domain
55 d2i6va1
not modelled
7.7
13
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like56 c3tdqB_
not modelled
7.4
20
PDB header: cell adhesionChain: B: PDB Molecule: pily2 protein;PDBTitle: crystal structure of a fimbrial biogenesis protein pily22 (pily2_pa4555) from pseudomonas aeruginosa pao1 at 2.10 a resolution
57 d1w0na_
not modelled
7.2
29
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 36 carbohydrate binding module, CBM3658 c2qtkB_
not modelled
7.0
10
PDB header: membrane proteinChain: B: PDB Molecule: probable porin;PDBTitle: crystal structure of the outer membrane protein opdk from2 pseudomonas aeruginosa
59 c2ihmA_
not modelled
6.9
14
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase mu;PDBTitle: polymerase mu in ternary complex with gapped 11mer dna2 duplex and bound incoming nucleotide
60 c1wx4B_
not modelled
6.7
28
PDB header: oxidoreductase/metal transportChain: B: PDB Molecule: melc;PDBTitle: crystal structure of the oxy-form of the copper-bound streptomyces2 castaneoglobisporus tyrosinase complexed with a caddie protein3 prepared by the addition of dithiothreitol
61 d2ef1a1
not modelled
6.6
8
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: ADP ribosyl cyclase-like62 d1isia_
not modelled
6.5
16
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: ADP ribosyl cyclase-like63 d1gm5a2
not modelled
6.2
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RecG "wedge" domain64 c8iczA_
not modelled
6.2
12
PDB header: transferase/dnaChain: A: PDB Molecule: protein (dna polymerase beta (e.c.2.7.7.7));PDBTitle: dna polymerase beta (pol b) (e.c.2.7.7.7) complexed with2 seven base pairs of dna; soaked in the presence of of datp3 (1 millimolar), mncl2 (5 millimolar), and lithium sulfate4 (75 millimolar)
65 c2y0lA_
not modelled
6.0
14
PDB header: transport proteinChain: A: PDB Molecule: cis-aconitate porin opdh;PDBTitle: crystal structure of pseudomonas aeruginosa opdo
66 d2aq0a1
not modelled
5.8
38
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like67 c3i18A_
not modelled
5.3
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2051 protein;PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
68 c2kjpA_
not modelled
5.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ylbl;PDBTitle: solution structure of protein ylbl (bsu15050) from bacillus2 subtilis, northeast structural genomics consortium target3 sr713a
69 d1qw2a_
not modelled
5.2
12
Fold: Hypothetical protein Ta1206Superfamily: Hypothetical protein Ta1206Family: Hypothetical protein Ta1206