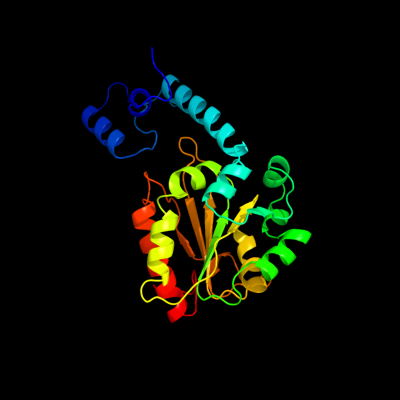
| 1 | c3absD_
|
|
 |
100.0 |
100 |
PDB header:lyase
Chain: D: PDB Molecule:ethanolamine ammonia-lyase light chain;
PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with adeninylpentylcobalamin and ethanolamine
|
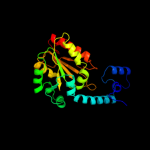
| 2 | c3anyB_
|
|
 |
100.0 |
100 |
PDB header:lyase
Chain: B: PDB Molecule:ethanolamine ammonia-lyase light chain;
PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with cn-cbl and (r)-2-amino-1-propanol
|
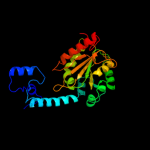
| 3 | c2e5cA_
|
|
 |
75.7 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinamide phosphoribosyltransferase;
PDBTitle: crystal structure of human nmprtase complexed with 5'-phosphoribosyl-2 1'-pyrophosphate
|
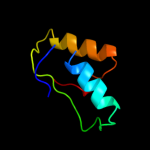
| 4 | d2f7fa1
|
|
 |
55.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 5 | c2i14B_
|
|
 |
51.5 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide2 pyrophosphorylase from pyrococcus furiosus
|
| 6 | d1yira1
|
|
 |
50.4 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
| 7 | d1dysa_
|
|
 |
49.6 |
18 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycosyl hydrolases family 6, cellulases
Family:Glycosyl hydrolases family 6, cellulases |
| 8 | c1yirA_
|
|
 |
46.1 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase 2;
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase
|
| 9 | d2huha1
|
|
 |
44.4 |
38 |
Fold:C2 domain-like
Superfamily:Smr-associated domain-like
Family:Smr-associated domain |
| 10 | d3bgsa1
|
|
 |
43.7 |
23 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 11 | d2i14a1
|
|
 |
43.4 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 12 | d3pnpa_
|
|
 |
42.0 |
23 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 13 | c3os4A_
|
|
 |
41.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: the crystal structure of nicotinate phosphoribosyltransferase from2 yersinia pestis
|
| 14 | d1ytda1
|
|
 |
36.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 15 | c3ggsA_
|
|
 |
36.3 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: human purine nucleoside phosphorylase double mutant e201q,n243d2 complexed with 2-fluoro-2'-deoxyadenosine
|
| 16 | c2p4sA_
|
|
 |
35.9 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
|
| 17 | d1qjwa_
|
|
 |
34.0 |
22 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycosyl hydrolases family 6, cellulases
Family:Glycosyl hydrolases family 6, cellulases |
| 18 | c3l0oB_
|
|
 |
33.8 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:transcription termination factor rho;
PDBTitle: structure of rna-free rho transcription termination factor from2 thermotoga maritima
|
| 19 | c2f7fA_
|
|
 |
32.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase, putative;
PDBTitle: crystal structure of enterococcus faecalis putative nicotinate2 phosphoribosyltransferase, new york structural genomics consortium
|
| 20 | d1ng6a_
|
|
 |
31.7 |
22 |
Fold:GatB/YqeY motif
Superfamily:GatB/YqeY motif
Family:GatB/YqeY domain |
| 21 | c1xpuB_ |
|
not modelled |
31.5 |
38 |
PDB header:transcription/rna
Chain: B: PDB Molecule:rho transcription termination factor;
PDBTitle: structural mechanism of inhibition of the rho transcription2 termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-3 dihydrobicyclomycin (fpdb)
|
| 22 | c3otbB_ |
|
not modelled |
31.2 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:trna(his) guanylyltransferase;
PDBTitle: crystal structure of human trnahis guanylyltransferase (thg1) - dgtp2 complex
|
| 23 | c2gsoB_ |
|
not modelled |
31.1 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:phosphodiesterase-nucleotide pyrophosphatase;
PDBTitle: structure of xac nucleotide2 pyrophosphatase/phosphodiesterase in complex with vanadate
|
| 24 | d2vapa1 |
|
not modelled |
29.4 |
19 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 25 | c1tcvB_ |
|
not modelled |
29.0 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:purine-nucleoside phosphorylase;
PDBTitle: crystal structure of the purine nucleoside phosphorylase2 from schistosoma mansoni in complex with non-detergent3 sulfobetaine 195 and acetate
|
| 26 | d1oc7a_ |
|
not modelled |
27.1 |
19 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycosyl hydrolases family 6, cellulases
Family:Glycosyl hydrolases family 6, cellulases |
| 27 | c3a64A_ |
|
not modelled |
25.2 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellobiohydrolase;
PDBTitle: crystal structure of cccel6c, a glycoside hydrolase family 62 enzyme, from coprinopsis cinerea
|
| 28 | d2jeka1 |
|
not modelled |
25.2 |
23 |
Fold:Rv1873-like
Superfamily:Rv1873-like
Family:Rv1873-like |
| 29 | d1dk0a_ |
|
not modelled |
24.7 |
25 |
Fold:Heme-binding protein A (HasA)
Superfamily:Heme-binding protein A (HasA)
Family:Heme-binding protein A (HasA) |
| 30 | d1ofua1 |
|
not modelled |
24.5 |
17 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 31 | c3la8A_ |
|
not modelled |
24.5 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:putative purine nucleoside phosphorylase;
PDBTitle: the crystal structure of smu.1229 from streptococcus mutans ua159
|
| 32 | d1qe5a_ |
|
not modelled |
23.9 |
15 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 33 | c3ddrC_ |
|
not modelled |
21.7 |
25 |
PDB header:membrane protein/heme binding protein
Chain: C: PDB Molecule:hemophore hasa;
PDBTitle: structure of the serratia marcescens hemophore receptor hasr-ile671gly2 mutant in complex with its hemophore hasa and heme
|
| 34 | d1gdta1 |
|
not modelled |
21.6 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 35 | c2zf8A_ |
|
not modelled |
21.4 |
24 |
PDB header:structural protein
Chain: A: PDB Molecule:component of sodium-driven polar flagellar motor;
PDBTitle: crystal structure of moty
|
| 36 | c1ybeA_ |
|
not modelled |
21.2 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase
|
| 37 | d1vmka_ |
|
not modelled |
20.8 |
15 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 38 | d1rvxa_ |
|
not modelled |
20.6 |
26 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 39 | c1vlpA_ |
|
not modelled |
19.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of a putative nicotinate phosphoribosyltransferase2 (yor209c, npt1) from saccharomyces cerevisiae at 1.75 a resolution
|
| 40 | c3dmaA_ |
|
not modelled |
19.1 |
5 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:exopolyphosphatase-related protein;
PDBTitle: crystal structure of an exopolyphosphatase-related protein2 from bacteroides fragilis. northeast structural genomics3 target bfr192
|
| 41 | c1ytkA_ |
|
not modelled |
19.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase from thermoplasma
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase from2 thermoplasma acidophilum with nicotinate mononucleotide
|
| 42 | c2rhoB_ |
|
not modelled |
18.8 |
19 |
PDB header:cell cycle
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: synthetic gene encoded bacillus subtilis ftsz ncs dimer with2 bound gdp and gtp-gamma-s
|
| 43 | c1w5fA_ |
|
not modelled |
18.6 |
17 |
PDB header:cell division
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
|
| 44 | c3crqA_ |
|
not modelled |
18.4 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: structure of trna dimethylallyltransferase: rna2 modification through a channel
|
| 45 | c1b0aA_ |
|
not modelled |
18.3 |
20 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
| 46 | c3szzA_ |
|
not modelled |
18.1 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonoacetate hydrolase;
PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
|
| 47 | c2p10D_ |
|
not modelled |
17.1 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 48 | c2c2pA_ |
|
not modelled |
16.8 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:g/u mismatch-specific dna glycosylase;
PDBTitle: the crystal structure of mismatch specific uracil-dna2 glycosylase (mug) from deinococcus radiodurans
|
| 49 | d1rq2a1 |
|
not modelled |
16.6 |
20 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 50 | d2p10a1 |
|
not modelled |
16.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
| 51 | c2vg2C_ |
|
not modelled |
16.1 |
25 |
PDB header:transferase
Chain: C: PDB Molecule:undecaprenyl pyrophosphate synthetase;
PDBTitle: rv2361 with ipp
|
| 52 | d1ybea1 |
|
not modelled |
15.7 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
| 53 | d1xpua3 |
|
not modelled |
15.5 |
27 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 54 | d2jdia3 |
|
not modelled |
15.5 |
10 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 55 | c2vawA_ |
|
not modelled |
15.2 |
20 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: ftsz pseudomonas aeruginosa gdp
|
| 56 | c3q3qA_ |
|
not modelled |
15.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase;
PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
|
| 57 | d1jlja_ |
|
not modelled |
14.6 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 58 | d1ns5a_ |
|
not modelled |
14.6 |
24 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 59 | d1w5fa1 |
|
not modelled |
14.3 |
17 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 60 | d1q6za1 |
|
not modelled |
14.3 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 61 | c3khsB_ |
|
not modelled |
14.1 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 62 | c2vxyA_ |
|
not modelled |
14.0 |
20 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: the structure of ftsz from bacillus subtilis at 1.7a2 resolution
|
| 63 | c2r6r1_ |
|
not modelled |
13.9 |
17 |
PDB header:cell cycle
Chain: 1: PDB Molecule:cell division protein ftsz;
PDBTitle: aquifex aeolicus ftsz
|
| 64 | c1e9kA_ |
|
not modelled |
13.8 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:camp specific phosphodiesterase pde4d5;
PDBTitle: the structure of the rack1 interaction sites located within2 the unique n-terminal region of the camp-specific3 phosphodiesterase, pde4d5.
|
| 65 | c1ofuB_ |
|
not modelled |
13.4 |
20 |
PDB header:bacterial cell division inhibitor
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of sula:ftsz from pseudomonas aeruginosa
|
| 66 | d1muga_ |
|
not modelled |
13.1 |
24 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:Mug-like |
| 67 | d2jdid3 |
|
not modelled |
13.0 |
50 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 68 | c2d3yA_ |
|
not modelled |
12.9 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: crystal structure of uracil-dna glycosylase from thermus thermophilus2 hb8
|
| 69 | c3ikbB_ |
|
not modelled |
12.9 |
56 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the structure of a conserved protein from streptococcus2 mutans ua159.
|
| 70 | d1fx0a3 |
|
not modelled |
12.9 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 71 | d1yo6a1 |
|
not modelled |
12.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | d1skye3 |
|
not modelled |
12.1 |
50 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 73 | c3cvjB_ |
|
not modelled |
11.8 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoheptose isomerase;
PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
|
| 74 | d1fx0b3 |
|
not modelled |
11.8 |
50 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 75 | c2rbaB_ |
|
not modelled |
11.6 |
24 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:g/t mismatch-specific thymine dna glycosylase;
PDBTitle: structure of human thymine dna glycosylase bound to abasic and2 undamaged dna
|
| 76 | c1w59B_ |
|
not modelled |
11.6 |
23 |
PDB header:cell division
Chain: B: PDB Molecule:cell division protein ftsz homolog 1;
PDBTitle: ftsz dimer, empty (m. jannaschii)
|
| 77 | c2c2xB_ |
|
not modelled |
11.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
|
| 78 | d1skyb3 |
|
not modelled |
11.4 |
8 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 79 | c2d07A_ |
|
not modelled |
11.1 |
39 |
PDB header:hydrolase
Chain: A: PDB Molecule:g/t mismatch-specific thymine dna glycosylase;
PDBTitle: crystal structure of sumo-3-modified thymine-dna glycosylase
|
| 80 | c3ragA_ |
|
not modelled |
10.5 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein aaci_0196 from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
|
| 81 | d1ecfa1 |
|
not modelled |
10.2 |
24 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 82 | d1d0gr1 |
|
not modelled |
10.1 |
45 |
Fold:TNF receptor-like
Superfamily:TNF receptor-like
Family:TNF receptor-like |
| 83 | c2l25A_ |
|
not modelled |
9.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: np_888769.1
|
| 84 | c3nglA_ |
|
not modelled |
9.8 |
20 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
|
| 85 | c3lxqB_ |
|
not modelled |
9.8 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein vp1736;
PDBTitle: the crystal structure of a protein in the alkaline2 phosphatase superfamily from vibrio parahaemolyticus to3 1.95a
|
| 86 | c3kzvA_ |
|
not modelled |
9.6 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized oxidoreductase yir035c;
PDBTitle: the crystal structure of a cytoplasmic protein with unknown function2 from saccharomyces cerevisiae
|
| 87 | d1fsua_ |
|
not modelled |
9.4 |
18 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 88 | c2q1yB_ |
|
not modelled |
9.4 |
19 |
PDB header:cell cycle, signaling protein
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of cell division protein ftsz from mycobacterium2 tuberculosis in complex with gtp-gamma-s
|
| 89 | c2l6pA_ |
|
not modelled |
9.3 |
18 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:phac1, phac2 and phad genes;
PDBTitle: nmr solution structure of the protein np_253742.1
|
| 90 | c1ecjB_ |
|
not modelled |
8.9 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:glutamine phosphoribosylpyrophosphate
PDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
|
| 91 | d1ei6a_ |
|
not modelled |
8.9 |
22 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Phosphonoacetate hydrolase |
| 92 | c3fybA_ |
|
not modelled |
8.7 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein of unknown function (duf1244);
PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
|
| 93 | d1kifa1 |
|
not modelled |
7.8 |
22 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:D-aminoacid oxidase, N-terminal domain |
| 94 | d1yt3a1 |
|
not modelled |
7.8 |
12 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:RNase D C-terminal domains |
| 95 | d1g2oa_ |
|
not modelled |
7.8 |
24 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 96 | c3h87D_ |
|
not modelled |
7.7 |
25 |
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
|
| 97 | d1wura1 |
|
not modelled |
7.5 |
10 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
| 98 | c1wm9D_ |
|
not modelled |
7.5 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: structure of gtp cyclohydrolase i from thermus thermophilus hb8
|
| 99 | d1hdha_ |
|
not modelled |
7.4 |
31 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |