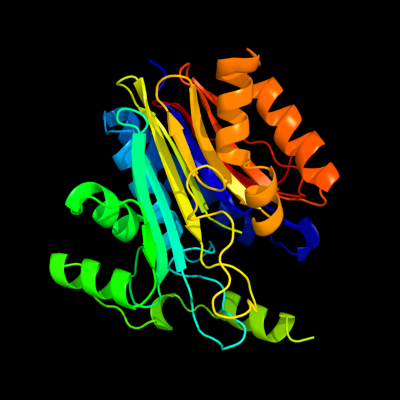
| 1 | c2zakB_
|
|
 |
100.0 |
99 |
PDB header:hydrolase
Chain: B: PDB Molecule:l-asparaginase precursor;
PDBTitle: orthorhombic crystal structure of precursor e. coli isoaspartyl2 peptidase/l-asparaginase (ecaiii) with active-site t179a mutation
|
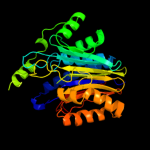
| 2 | c1p4vA_
|
|
 |
100.0 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:n(4)-(beta-n-acetylglucosaminyl)-l-asparaginase
PDBTitle: crystal structure of the glycosylasparaginase precursor2 d151n mutant with glycine
|
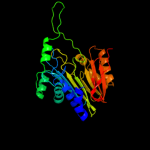
| 3 | c2a8lB_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:threonine aspartase 1;
PDBTitle: crystal structure of human taspase1 (t234a mutant)
|
| 4 | c1t3mA_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative l-asparaginase;
PDBTitle: structure of the isoaspartyl peptidase with l-asparaginase2 activity from e. coli
|
| 5 | c2gezE_
|
|
 |
100.0 |
40 |
PDB header:hydrolase
Chain: E: PDB Molecule:l-asparaginase alpha subunit;
PDBTitle: crystal structure of potassium-independent plant asparaginase
|
| 6 | c2zalD_
|
|
 |
100.0 |
99 |
PDB header:hydrolase
Chain: D: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of e. coli isoaspartyl aminopeptidase/l-asparaginase2 in complex with l-aspartate
|
| 7 | c2zalB_
|
|
 |
100.0 |
99 |
PDB header:hydrolase
Chain: B: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of e. coli isoaspartyl aminopeptidase/l-asparaginase2 in complex with l-aspartate
|
| 8 | c1k2xB_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative l-asparaginase;
PDBTitle: crystal structure of putative asparaginase encoded by escherichia coli2 ybik gene
|
| 9 | c1k2xD_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative l-asparaginase;
PDBTitle: crystal structure of putative asparaginase encoded by escherichia coli2 ybik gene
|
| 10 | c1jn9D_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative l-asparaginase;
PDBTitle: structure of putative asparaginase encoded by escherichia coli ybik2 gene
|
| 11 | c1t3mD_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative l-asparaginase;
PDBTitle: structure of the isoaspartyl peptidase with l-asparaginase2 activity from e. coli
|
| 12 | c1jn9B_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative l-asparaginase;
PDBTitle: structure of putative asparaginase encoded by escherichia coli ybik2 gene
|
| 13 | c1t3mB_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative l-asparaginase;
PDBTitle: structure of the isoaspartyl peptidase with l-asparaginase2 activity from e. coli
|
| 14 | c1apyA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:aspartylglucosaminidase;
PDBTitle: human aspartylglucosaminidase
|
| 15 | c2gezF_
|
|
 |
100.0 |
55 |
PDB header:hydrolase
Chain: F: PDB Molecule:l-asparaginase beta subunit;
PDBTitle: crystal structure of potassium-independent plant asparaginase
|
| 16 | c2gacA_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosylasparaginase;
PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
|
| 17 | c2gacD_
|
|
 |
100.0 |
34 |
PDB header:hydrolase
Chain: D: PDB Molecule:glycosylasparaginase;
PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
|
| 18 | c1apzB_
|
|
 |
100.0 |
32 |
PDB header:complex (hydrolase/peptide)
Chain: B: PDB Molecule:aspartylglucosaminidase;
PDBTitle: human aspartylglucosaminidase complex with reaction product
|
| 19 | c2e0wA_
|
|
 |
97.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: t391a precursor mutant protein of gamma-glutamyltranspeptidase from2 escherichia coli
|
| 20 | c2e0yB_
|
|
 |
95.0 |
37 |
PDB header:transferase
Chain: B: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: crystal structure of the samarium derivative of mature gamma-2 glutamyltranspeptidase from escherichia coli
|
| 21 | d2nlza1 |
|
not modelled |
94.6 |
26 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Gamma-glutamyltranspeptidase-like |
| 22 | c2z8jA_ |
|
not modelled |
94.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: crystal structure of escherichia coli gamma-2 glutamyltranspeptidase in complex with azaserine prepared3 in the dark
|
| 23 | c2qm6C_ |
|
not modelled |
93.3 |
32 |
PDB header:transferase
Chain: C: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase2 in complex with glutamate
|
| 24 | c2v36A_ |
|
not modelled |
92.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyltranspeptidase large chain;
PDBTitle: crystal structure of gamma-glutamyl transferase from2 bacillus subtilis
|
| 25 | c3g9kD_ |
|
not modelled |
92.8 |
35 |
PDB header:hydrolase
Chain: D: PDB Molecule:capsule biosynthesis protein capd;
PDBTitle: crystal structure of bacillus anthracis transpeptidase enzyme capd
|
| 26 | d2i3oa1 |
|
not modelled |
92.6 |
14 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Gamma-glutamyltranspeptidase-like |
| 27 | c3ga9S_ |
|
not modelled |
91.9 |
24 |
PDB header:hydrolase
Chain: S: PDB Molecule:capsule biosynthesis protein capd;
PDBTitle: crystal structure of bacillus anthracis transpeptidase enzyme capd,2 crystal form ii
|
| 28 | d2imha1 |
|
not modelled |
91.7 |
20 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:SPO2555-like |
| 29 | c2v36D_ |
|
not modelled |
88.3 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:gamma-glutamyltranspeptidase small chain;
PDBTitle: crystal structure of gamma-glutamyl transferase from2 bacillus subtilis
|
| 30 | c2nqoB_ |
|
not modelled |
78.9 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase
|
| 31 | d1k4ia_ |
|
not modelled |
48.1 |
17 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 32 | c2otdC_ |
|
not modelled |
31.1 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:glycerophosphodiester phosphodiesterase;
PDBTitle: the crystal structure of the glycerophosphodiester phosphodiesterase2 from shigella flexneri 2a
|
| 33 | d1hw8a2 |
|
not modelled |
23.7 |
27 |
Fold:Substrate-binding domain of HMG-CoA reductase
Superfamily:Substrate-binding domain of HMG-CoA reductase
Family:Substrate-binding domain of HMG-CoA reductase |
| 34 | c3mioA_ |
|
not modelled |
23.2 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:3,4-dihydroxy-2-butanone 4-phosphate synthase;
PDBTitle: crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase2 domain from mycobacterium tuberculosis at ph 6.00
|
| 35 | c1yy3A_ |
|
not modelled |
21.5 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:s-adenosylmethionine:trna ribosyltransferase-
PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
|
| 36 | d1tqha_ |
|
not modelled |
21.0 |
22 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase/lipase |
| 37 | c3cdkD_ |
|
not modelled |
19.0 |
32 |
PDB header:transferase
Chain: D: PDB Molecule:succinyl-coa:3-ketoacid-coenzyme a transferase
PDBTitle: crystal structure of the co-expressed succinyl-coa2 transferase a and b complex from bacillus subtilis
|
| 38 | d1tksa_ |
|
not modelled |
19.0 |
11 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 39 | d1g57a_ |
|
not modelled |
18.5 |
19 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 40 | d2nn6h2 |
|
not modelled |
16.6 |
20 |
Fold:Barrel-sandwich hybrid
Superfamily:Ribosomal L27 protein-like
Family:ECR1 N-terminal domain-like |
| 41 | d2djia1 |
|
not modelled |
16.6 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 42 | d1k25a4 |
|
not modelled |
16.2 |
13 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 43 | c3m1eA_ |
|
not modelled |
16.1 |
18 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: crystal structure of benm_dbd
|
| 44 | d2ihta1 |
|
not modelled |
15.8 |
25 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 45 | d1azwa_ |
|
not modelled |
15.4 |
54 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Proline iminopeptidase-like |
| 46 | d1t9ba1 |
|
not modelled |
15.3 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 47 | d1dqaa4 |
|
not modelled |
15.2 |
27 |
Fold:Substrate-binding domain of HMG-CoA reductase
Superfamily:Substrate-binding domain of HMG-CoA reductase
Family:Substrate-binding domain of HMG-CoA reductase |
| 48 | d2phna1 |
|
not modelled |
14.8 |
25 |
Fold:CofE-like
Superfamily:CofE-like
Family:CofE-like |
| 49 | d1kyqa2 |
|
not modelled |
14.8 |
30 |
Fold:Siroheme synthase middle domains-like
Superfamily:Siroheme synthase middle domains-like
Family:Siroheme synthase middle domains-like |
| 50 | c2qruA_ |
|
not modelled |
14.7 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an alpha/beta hydrolase superfamily protein from2 enterococcus faecalis
|
| 51 | c1qgeD_ |
|
not modelled |
14.6 |
46 |
PDB header:hydrolase
Chain: D: PDB Molecule:protein (triacylglycerol hydrolase);
PDBTitle: new crystal form of pseudomonas glumae (formerly chromobacterium2 viscosum atcc 6918) lipase
|
| 52 | c3hxkB_ |
|
not modelled |
14.5 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:sugar hydrolase;
PDBTitle: crystal structure of a sugar hydrolase (yeeb) from2 lactococcus lactis, northeast structural genomics3 consortium target kr108
|
| 53 | c3fnbB_ |
|
not modelled |
14.3 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:acylaminoacyl peptidase smu_737;
PDBTitle: crystal structure of acylaminoacyl peptidase smu_737 from2 streptococcus mutans ua159
|
| 54 | d1n9ba_ |
|
not modelled |
14.0 |
19 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 55 | c3ms6A_ |
|
not modelled |
13.9 |
27 |
PDB header:protein binding
Chain: A: PDB Molecule:hepatitis b virus x-interacting protein;
PDBTitle: crystal structure of hepatitis b x-interacting protein (hbxip)
|
| 56 | d1zpda1 |
|
not modelled |
13.8 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 57 | d2nn6i2 |
|
not modelled |
13.8 |
25 |
Fold:Barrel-sandwich hybrid
Superfamily:Ribosomal L27 protein-like
Family:ECR1 N-terminal domain-like |
| 58 | d1hw8c2 |
|
not modelled |
13.7 |
27 |
Fold:Substrate-binding domain of HMG-CoA reductase
Superfamily:Substrate-binding domain of HMG-CoA reductase
Family:Substrate-binding domain of HMG-CoA reductase |
| 59 | c3bitA_ |
|
not modelled |
13.6 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: crystal structure of yeast spt16 n-terminal domain
|
| 60 | d2ez9a1 |
|
not modelled |
13.5 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 61 | d1wjsa_ |
|
not modelled |
13.3 |
13 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 62 | c3ue3A_ |
|
not modelled |
13.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:septum formation, penicillin binding protein 3,
PDBTitle: crystal structure of acinetobacter baumanni pbp3
|
| 63 | c2op8A_ |
|
not modelled |
13.2 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase ywhb;
PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
|
| 64 | c2gzqA_ |
|
not modelled |
13.0 |
12 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:phosphatidylethanolamine-binding protein;
PDBTitle: phosphatidylethanolamine-binding protein from plasmodium vivax
|
| 65 | c1kyqC_ |
|
not modelled |
12.9 |
28 |
PDB header:oxidoreductase, lyase
Chain: C: PDB Molecule:siroheme biosynthesis protein met8;
PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
|
| 66 | c1w7vD_ |
|
not modelled |
12.9 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:xaa-pro aminopeptidase;
PDBTitle: znmg substituted aminopeptidase p from e. coli
|
| 67 | c1alnA_ |
|
not modelled |
12.6 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytidine deaminase;
PDBTitle: crystal structure of cytidine deaminase complexed with 3-deazacytidine
|
| 68 | d1qlwa_ |
|
not modelled |
12.5 |
27 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:A novel bacterial esterase |
| 69 | c1hwjB_ |
|
not modelled |
12.3 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:hmg-coa reductase;
PDBTitle: complex of the catalytic portion of human hmg-coa reductase2 with cerivastatin
|
| 70 | c3dclC_ |
|
not modelled |
12.2 |
27 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:tm1086;
PDBTitle: crystal structure of tm1086
|
| 71 | c3cd0B_ |
|
not modelled |
12.2 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxy-3-methylglutaryl-coenzyme a reductase;
PDBTitle: thermodynamic and structure guided design of statin hmg-coa2 reductase inhibitors
|
| 72 | d1pvda1 |
|
not modelled |
12.1 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 73 | d2ji7a1 |
|
not modelled |
12.0 |
34 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 74 | c2l09A_ |
|
not modelled |
12.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:asr4154 protein;
PDBTitle: solution nmr structure of protein asr4154 from nostoc sp. pcc71202 northeast structural genomics consortium target id nsr143
|
| 75 | d2bgra2 |
|
not modelled |
11.8 |
9 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:DPP6 catalytic domain-like |
| 76 | d1dosa_ |
|
not modelled |
11.7 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 77 | c3cb5A_ |
|
not modelled |
11.7 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: crystal structure of the s. pombe peptidase homology domain of fact2 complex subunit spt16 (form a)
|
| 78 | c3h04A_ |
|
not modelled |
11.4 |
50 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the protein with unknown function from2 staphylococcus aureus subsp. aureus mu50
|
| 79 | c2x4kB_ |
|
not modelled |
11.3 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
|
| 80 | d1bjpa_ |
|
not modelled |
11.0 |
23 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
| 81 | c2g6pA_ |
|
not modelled |
11.0 |
2 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase 1;
PDBTitle: crystal structure of truncated (delta 1-89) human methionine2 aminopeptidase type 1 in complex with pyridyl pyrimidine derivative
|
| 82 | c2gz5A_ |
|
not modelled |
11.0 |
2 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase 1;
PDBTitle: human type 1 methionine aminopeptidase in complex with ovalicin at 1.12 ang
|
| 83 | d1alna1 |
|
not modelled |
10.7 |
19 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
| 84 | d1pjaa_ |
|
not modelled |
10.6 |
42 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Thioesterases |
| 85 | c1pjaA_ |
|
not modelled |
10.6 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:palmitoyl-protein thioesterase 2 precursor;
PDBTitle: the crystal structure of palmitoyl protein thioesterase-2 reveals the2 basis for divergent substrate specificities of the two lysosomal3 thioesterases (ppt1 and ppt2)
|
| 86 | d1snna_ |
|
not modelled |
10.4 |
16 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 87 | c3oc2A_ |
|
not modelled |
10.3 |
23 |
PDB header:penicillin-binding protein
Chain: A: PDB Molecule:penicillin-binding protein 3;
PDBTitle: crystal structure of penicillin-binding protein 3 from pseudomonas2 aeruginosa
|
| 88 | d1oxwa_ |
|
not modelled |
10.3 |
45 |
Fold:FabD/lysophospholipase-like
Superfamily:FabD/lysophospholipase-like
Family:Patatin |
| 89 | d2pmra1 |
|
not modelled |
10.2 |
14 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:AF1782-like
Family:AF1782-like |
| 90 | c3llcA_ |
|
not modelled |
10.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of putative hydrolase (yp_002548124.1) from2 agrobacterium vitis s4 at 1.80 a resolution
|
| 91 | d1q6za1 |
|
not modelled |
9.9 |
9 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 92 | c2jz5A_ |
|
not modelled |
9.8 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vpa0419;
PDBTitle: nmr solution structure of protein vpa0419 from vibrio2 parahaemolyticus. northeast structural genomics target3 vpr68
|
| 93 | c3dgsA_ |
|
not modelled |
9.6 |
31 |
PDB header:viral protein
Chain: A: PDB Molecule:coat protein a;
PDBTitle: changing the determinants of protein stability from2 covalent to non-covalent interactions by in-vitro3 evolution: a structural and energetic analysis
|
| 94 | c1chmA_ |
|
not modelled |
9.6 |
22 |
PDB header:creatinase
Chain: A: PDB Molecule:creatine amidinohydrolase;
PDBTitle: enzymatic mechanism of creatine amidinohydrolase as deduced2 from crystal structures
|
| 95 | c2kdvA_ |
|
not modelled |
9.6 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:rna pyrophosphohydrolase;
PDBTitle: solution structure of rna pyrophosphohydrolase rpph from2 escherichia coli
|
| 96 | d2nn6c2 |
|
not modelled |
9.6 |
17 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 97 | c3gy9A_ |
|
not modelled |
9.6 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:gcn5-related n-acetyltransferase;
PDBTitle: crystal structure of putative acetyltransferase (yp_001815201.1) from2 exiguobacterium sp. 255-15 at 1.52 a resolution
|
| 98 | c2z0rA_ |
|
not modelled |
9.5 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized protein ttha0547;
PDBTitle: crystal structure of hypothetical protein ttha0547
|
| 99 | d1xkla_ |
|
not modelled |
9.5 |
31 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Hydroxynitrile lyase-like |