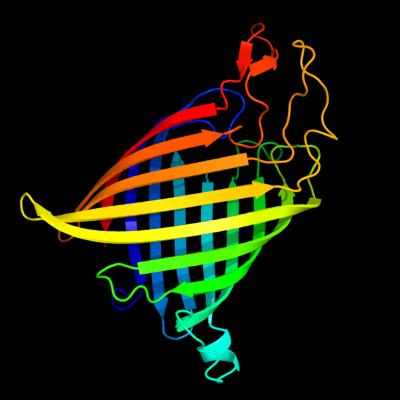
1 d1tlya_
100.0
100
Fold: Transmembrane beta-barrelsSuperfamily: Tsx-like channelFamily: Tsx-like channel2 d2e9xb2
20.4
41
Fold: GINS/PriA/YqbF domainSuperfamily: PriA/YqbF domainFamily: PSF2 N-terminal domain-like3 d2jnaa1
20.3
39
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like4 c3epzA_
20.2
33
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: structure of the replication foci-targeting sequence of human dna2 cytosine methyltransferase dnmt1
5 c3f09B_
18.9
27
PDB header: transferaseChain: B: PDB Molecule: holo-[acyl-carrier-protein] synthase;PDBTitle: 1.82 angstrom resolution crystal structure of holo-(acyl-carrier-2 protein) synthase (acps) from staphylococcus aureus
6 c1u8cB_
17.7
16
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
7 c3n6rK_
16.7
25
PDB header: ligaseChain: K: PDB Molecule: propionyl-coa carboxylase, alpha subunit;PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
8 c2e9xF_
15.5
41
PDB header: replicationChain: F: PDB Molecule: dna replication complex gins protein psf2;PDBTitle: the crystal structure of human gins core complex
9 d2j9ga3
14.0
29
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like10 c3u9sE_
13.7
33
PDB header: ligaseChain: E: PDB Molecule: methylcrotonyl-coa carboxylase, alpha-subunit;PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
11 c2x27X_
13.1
17
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
12 c3g8cB_
12.5
27
PDB header: ligaseChain: B: PDB Molecule: biotin carboxylase;PDBTitle: crystal stucture of biotin carboxylase in complex with2 biotin, bicarbonate, adp and mg ion
13 c2k3aA_
12.5
15
PDB header: hydrolaseChain: A: PDB Molecule: chap domain protein;PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
14 d1w96a3
12.0
24
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like15 d1gsoa3
10.0
35
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like16 c2qdzA_
9.3
17
PDB header: protein transportChain: A: PDB Molecule: tpsb transporter fhac;PDBTitle: structure of the membrane protein fhac: a member of the2 omp85/tpsb transporter family
17 c2vpqA_
7.6
32
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase;PDBTitle: crystal structure of biotin carboxylase from s. aureus2 complexed with amppnp
18 c3bg5C_
7.6
25
PDB header: ligaseChain: C: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of staphylococcus aureus pyruvate2 carboxylase
19 c2pvpB_
7.5
21
PDB header: ligaseChain: B: PDB Molecule: d-alanine-d-alanine ligase;PDBTitle: crystal structure of d-alanine-d-alanine ligase from helicobacter2 pylori
20 c2gpwC_
7.3
27
PDB header: ligaseChain: C: PDB Molecule: biotin carboxylase;PDBTitle: crystal structure of the biotin carboxylase subunit, f363a2 mutant, of acetyl-coa carboxylase from escherichia coli.
21 c2hjwA_
not modelled
7.2
29
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: crystal structure of the bc domain of acc2
22 c1w96B_
not modelled
7.0
24
PDB header: ligaseChain: B: PDB Molecule: acetyl-coenzyme a carboxylase;PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
23 d1iu4a_
not modelled
6.5
21
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Microbial transglutaminase24 d1ev11_
not modelled
6.4
19
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Picornaviridae-like VP (VP1, VP2, VP3 and VP4)25 c3iu0A_
not modelled
6.1
21
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: structural basis for zymogen activation and substrate binding of2 transglutaminase from streptomyces mobaraense
26 c3lyvF_
not modelled
5.8
24
PDB header: chaperoneChain: F: PDB Molecule: ribosome-associated factor y;PDBTitle: crystal structure of a domain of ribosome-associated factor y from2 streptococcus pyogenes serotype m6. northeast structural genomics3 consortium target id dr64a
27 c3k6sB_
not modelled
5.8
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
28 c2ktlA_
not modelled
5.7
30
PDB header: ligaseChain: A: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: structure of c-terminal domain from mttyrrs of a. nidulans
29 d1vkza3
not modelled
5.6
25
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like30 c2k0lA_
not modelled
5.6
12
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
31 c3ouzA_
not modelled
5.5
15
PDB header: ligaseChain: A: PDB Molecule: biotin carboxylase;PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
32 c2q5xA_
not modelled
5.5
29
PDB header: protein transportChain: A: PDB Molecule: nuclear pore complex protein nup98;PDBTitle: crystal structure of the c-terminal domain of hnup98
33 c2jmmA_
not modelled
5.3
20
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
34 d1jyaa_
not modelled
5.2
19
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Type III secretory system chaperone