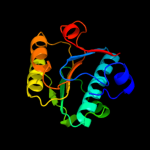
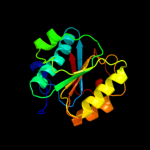
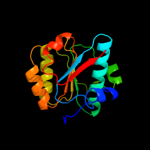
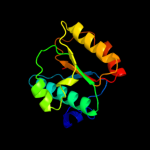
1 c1oftC_
100.0
26
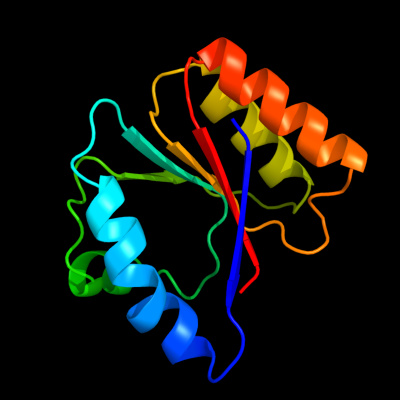
PDB header: bacterial cell division inhibitorChain: C: PDB Molecule: hypothetical protein pa3008;PDBTitle: crystal structure of sula from pseudomonas aeruginosa
2 d1ofux_
100.0
27
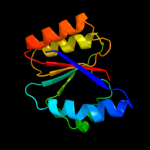
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Bacterial cell division inhibitor SulA3 d1xp8a1
99.5
16
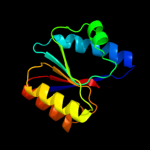
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)4 c1xp8A_
99.5
16
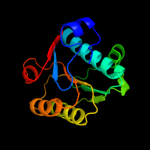
PDB header: dna binding proteinChain: A: PDB Molecule: reca protein;PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
5 d1ubea1
99.4
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)6 d1u94a1
99.4
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)7 c2zroA_
99.2
11
PDB header: hydrolaseChain: A: PDB Molecule: protein reca;PDBTitle: msreca adp form iv
8 c3hr8A_
99.2
14
PDB header: recombinationChain: A: PDB Molecule: protein reca;PDBTitle: crystal structure of thermotoga maritima reca
9 c2recB_
99.2
15
PDB header: helicasePDB COMPND: 10 d1mo6a1
99.1
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)11 c3cmwA_
98.9
13
PDB header: recombination/dnaChain: A: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
12 c3cmvG_
98.1
13
PDB header: recombinationChain: G: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
13 c3io5B_
97.9
14
PDB header: dna binding proteinChain: B: PDB Molecule: recombination and repair protein;PDBTitle: crystal structure of a dimeric form of the uvsx recombinase core2 domain from enterobacteria phage t4
14 c3cmuA_
97.4
16
PDB header: recombination/dnaChain: A: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
15 d1tf7a2
72.1
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)16 c2w0mA_
60.4
10
PDB header: unknown functionChain: A: PDB Molecule: sso2452;PDBTitle: crystal structure of sso2452 from sulfolobus solfataricus2 p2
17 c2ztsB_
59.7
11
PDB header: atp-binding proteinChain: B: PDB Molecule: putative uncharacterized protein ph0186;PDBTitle: crystal structure of kaic-like protein ph0186 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
18 d1j5pa4
58.4
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain19 c2dc1A_
55.9
7
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
20 c3ldaA_
40.8
18
PDB header: dna binding proteinChain: A: PDB Molecule: dna repair protein rad51;PDBTitle: yeast rad51 h352y filament interface mutant
21 d1rvv1_
not modelled
37.4
10
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase22 c2pjkA_
not modelled
37.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 178aa long hypothetical molybdenum cofactorPDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
23 d1nqua_
not modelled
33.5
10
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase24 c2obxH_
not modelled
33.3
17
PDB header: transferaseChain: H: PDB Molecule: 6,7-dimethyl-8-ribityllumazine synthase 1;PDBTitle: lumazine synthase ribh2 from mesorhizobium loti (gene mll7281, swiss-2 prot entry q986n2) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
25 c1t4gA_
not modelled
31.0
11
PDB header: recombinationChain: A: PDB Molecule: dna repair and recombination protein rada;PDBTitle: atpase in complex with amp-pnp
26 d1di0a_
not modelled
31.0
16
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase27 c2de4B_
not modelled
30.5
17
PDB header: hydrolaseChain: B: PDB Molecule: dibenzothiophene desulfurization enzyme b;PDBTitle: crystal structure of dszb c27s mutant in complex with biphenyl-2-2 sulfinic acid
28 c2dr3A_
not modelled
27.9
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0273 protein ph0284;PDBTitle: crystal structure of reca superfamily atpase ph0284 from pyrococcus2 horikoshii ot3
29 c1j5pA_
not modelled
27.3
15
PDB header: oxidoreductaseChain: A: PDB Molecule: aspartate dehydrogenase;PDBTitle: crystal structure of aspartate dehydrogenase (tm1643) from thermotoga2 maritima at 1.9 a resolution
30 c3k2gA_
not modelled
25.6
12
PDB header: resiniferatoxin binding proteinChain: A: PDB Molecule: resiniferatoxin-binding, phosphotriesterase-PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
31 c2is8A_
not modelled
24.1
7
PDB header: structural proteinChain: A: PDB Molecule: molybdopterin biosynthesis enzyme, moab;PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
32 d1y5ea1
not modelled
23.5
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like33 d1n0wa_
not modelled
22.7
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)34 d1szpa2
not modelled
21.9
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)35 c2i4cA_
not modelled
21.7
19
PDB header: transport proteinChain: A: PDB Molecule: bicarbonate transporter;PDBTitle: crystal structure of bicarbonate transport protein cmpa from2 synechocystis sp. pcc 6803 in complex with bicarbonate and calcium
36 d1srva_
not modelled
20.9
37
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain37 d1dk7a_
not modelled
19.8
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain38 d1ydwa1
not modelled
18.0
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain39 d2i1qa2
not modelled
17.9
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)40 c2wv9A_
not modelled
17.5
14
PDB header: hydrolaseChain: A: PDB Molecule: flavivirin protease ns2b regulatory subunit, flavivirinPDBTitle: crystal structure of the ns3 protease-helicase from murray2 valley encephalitis virus
41 d1v5wa_
not modelled
16.0
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)42 d1c2ya_
not modelled
15.7
5
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase43 d1we3a2
not modelled
15.5
37
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain44 d1oela2
not modelled
15.4
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain45 d1sjpa2
not modelled
15.3
32
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain46 d7aata_
not modelled
14.0
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like47 d1kida_
not modelled
13.9
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain48 c3gr1A_
not modelled
13.6
9
PDB header: membrane proteinChain: A: PDB Molecule: protein prgh;PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
49 d1o6za1
not modelled
13.0
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like50 d1i0za1
not modelled
12.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like51 d1o4sa_
not modelled
12.2
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like52 d1tf7a1
not modelled
12.1
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)53 d2ay1a_
not modelled
12.0
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like54 d1ldma1
not modelled
11.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like55 c3m6cA_
not modelled
11.9
32
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 1;PDBTitle: crystal structure of mycobacterium tuberculosis groel1 apical domain
56 c3mk3L_
not modelled
11.3
10
PDB header: transferaseChain: L: PDB Molecule: 6,7-dimethyl-8-ribityllumazine synthase;PDBTitle: crystal structure of lumazine synthase from salmonella typhimurium lt2
57 c3oirA_
not modelled
11.2
7
PDB header: transport proteinChain: A: PDB Molecule: sulfate transporter sulfate transporter family protein;PDBTitle: crystal structure of sulfate transporter family protein from wolinella2 succinogenes
58 c1ofgF_
not modelled
10.9
20
PDB header: oxidoreductaseChain: F: PDB Molecule: glucose-fructose oxidoreductase;PDBTitle: glucose-fructose oxidoreductase
59 d1c7na_
not modelled
10.7
6
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like60 d1bw0a_
not modelled
10.5
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like61 d2q7wa1
not modelled
10.4
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like62 c2dflA_
not modelled
10.4
13
PDB header: recombinationChain: A: PDB Molecule: dna repair and recombination protein rada;PDBTitle: crystal structure of left-handed rada filament
63 c3o82B_
not modelled
10.3
11
PDB header: ligaseChain: B: PDB Molecule: peptide arylation enzyme;PDBTitle: structure of base n-terminal domain from acinetobacter baumannii bound2 to 5'-o-[n-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine
64 c3ix1A_
not modelled
10.1
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
65 c3ix1B_
not modelled
10.1
18
PDB header: biosynthetic proteinChain: B: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
66 c3un6A_
not modelled
9.5
10
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein saouhsc_00137;PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
67 d1ebfa1
not modelled
9.2
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain68 d1ldna1
not modelled
9.1
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like69 d1mkza_
not modelled
9.1
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like70 c3btuD_
not modelled
8.9
14
PDB header: transcriptionChain: D: PDB Molecule: galactose/lactose metabolism regulatory proteinPDBTitle: crystal structure of the super-repressor mutant of gal80p2 from saccharomyces cerevisiae; gal80(s2) [e351k]
71 c1szpC_
not modelled
8.6
14
PDB header: dna binding proteinChain: C: PDB Molecule: dna repair protein rad51;PDBTitle: a crystal structure of the rad51 filament
72 c3f4lF_
not modelled
8.4
19
PDB header: oxidoreductaseChain: F: PDB Molecule: putative oxidoreductase yhhx;PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
73 d2nu7a1
not modelled
8.3
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain74 d1lc0a1
not modelled
8.2
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain75 d1m5wa_
not modelled
8.2
13
Fold: TIM beta/alpha-barrelSuperfamily: Pyridoxine 5'-phosphate synthaseFamily: Pyridoxine 5'-phosphate synthase76 c1h6dL_
not modelled
7.9
18
PDB header: protein translocationChain: L: PDB Molecule: precursor form of glucose-fructosePDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
77 c3gr0D_
not modelled
7.7
9
PDB header: membrane proteinChain: D: PDB Molecule: protein prgh;PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
78 d1np7a2
not modelled
7.6
29
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain79 c3kxwA_
not modelled
7.6
12
PDB header: ligaseChain: A: PDB Molecule: saframycin mx1 synthetase b;PDBTitle: the crystal structure of fatty acid amp ligase from legionella2 pneumophila
80 d2j07a2
not modelled
7.5
24
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain81 c3r4rB_
not modelled
7.4
25
PDB header: cell adhesionChain: B: PDB Molecule: hypothetical fimbrial assembly protein;PDBTitle: crystal structure of a hypothetical fimbrial assembly protein2 (bdi_3522) from parabacteroides distasonis atcc 8503 at 2.38 a3 resolution
82 c2x7pA_
not modelled
7.2
7
PDB header: unknown functionChain: A: PDB Molecule: possible thiamine biosynthesis enzyme;PDBTitle: the conserved candida albicans ca3427 gene product defines a new2 family of proteins exhibiting the generic periplasmic binding3 protein structural fold
83 c3dtyA_
not modelled
7.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, gfo/idh/moca family;PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
84 c3e2yB_
not modelled
6.9
12
PDB header: transferase, lyaseChain: B: PDB Molecule: kynurenine-oxoglutarate transaminase 3;PDBTitle: crystal structure of mouse kynurenine aminotransferase iii in complex2 with glutamine
85 d1owla2
not modelled
6.9
16
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain86 d2gdta1
not modelled
6.7
18
Fold: SARS Nsp1-likeSuperfamily: SARS Nsp1-likeFamily: SARS Nsp1-like87 d1xi9a_
not modelled
6.7
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like88 c3bgwD_
not modelled
6.6
8
PDB header: replicationChain: D: PDB Molecule: dnab-like replicative helicase;PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
89 d1wu2a3
not modelled
6.5
10
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like90 c2g29A_
not modelled
6.3
19
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein nrta;PDBTitle: crystal structure of the periplasmic nitrate-binding2 protein nrta from synechocystis pcc 6803
91 c3fd8A_
not modelled
6.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, gfo/idh/moca family;PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
92 c3umvB_
not modelled
6.3
22
PDB header: lyaseChain: B: PDB Molecule: deoxyribodipyrimidine photo-lyase;PDBTitle: eukaryotic class ii cpd photolyase structure reveals a basis for2 improved uv-tolerance in plants
93 d1to3a_
not modelled
6.1
11
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase94 d1uxja1
not modelled
6.1
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like95 d1y6ja1
not modelled
6.1
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like96 c3r0zA_
not modelled
5.5
22
PDB header: lyaseChain: A: PDB Molecule: d-serine dehydratase;PDBTitle: crystal structure of apo d-serine deaminase from salmonella2 typhimurium
97 d1guza1
not modelled
5.5
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like98 c3ahhA_
not modelled
5.5
7
PDB header: lyaseChain: A: PDB Molecule: xylulose 5-phosphate/fructose 6-phosphate phosphoketolase;PDBTitle: h142a mutant of phosphoketolase from bifidobacterium breve complexed2 with acetyl thiamine diphosphate
99 c3kbqA_
not modelled
5.5
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein ta0487;PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum