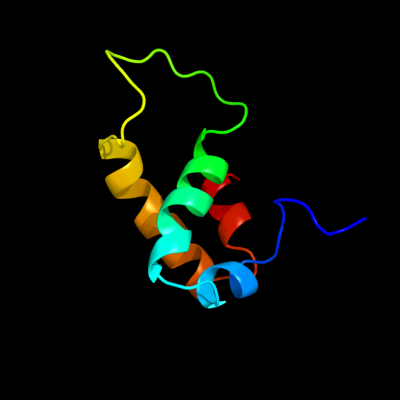
1 c2xuvB_
100.0
90
PDB header: unknown functionChain: B: PDB Molecule: hdeb;PDBTitle: the structure of hdeb
2 d1dj8a_
96.5
18
Fold: Protein HNS-dependent expression A; HdeASuperfamily: Protein HNS-dependent expression A; HdeAFamily: Protein HNS-dependent expression A; HdeA3 d2dk8a1
29.9
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RPO3F domain-like4 d1fs1b2
29.2
32
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain5 d1t3wa_
26.3
9
Fold: DNA primase DnaG, C-terminal domainSuperfamily: DNA primase DnaG, C-terminal domainFamily: DNA primase DnaG, C-terminal domain6 d2ezwa1
19.6
9
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit7 d2pi2e1
18.4
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB8 c3kwrA_
17.1
18
PDB header: rna binding proteinChain: A: PDB Molecule: putative rna-binding protein;PDBTitle: crystal structure of putative rna-binding protein (np_785364.1) from2 lactobacillus plantarum at 1.45 a resolution
9 c2pqaB_
16.5
14
PDB header: replicationChain: B: PDB Molecule: replication protein a 14 kda subunit;PDBTitle: crystal structure of full-length human rpa 14/32 heterodimer
10 d1nexa2
16.3
19
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain11 d1hv2a_
12.9
23
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain12 c2dadA_
12.8
20
PDB header: oncoproteinChain: A: PDB Molecule: absent in melanoma 1 protein;PDBTitle: solution structure of the fifth crystall domain of the non-2 lens protein, absent in melanoma 1
13 c3iuwA_
12.1
20
PDB header: rna binding proteinChain: A: PDB Molecule: activating signal cointegrator;PDBTitle: crystal structure of activating signal cointegrator (np_814290.1) from2 enterococcus faecalis v583 at 1.58 a resolution
14 c2kddB_
10.7
28
PDB header: cell cycleChain: B: PDB Molecule: borealin;PDBTitle: solution structure of the conserved c-terminal dimerization2 domain of borealin
15 d1vqoq1
9.2
20
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e16 c3iz6M_
9.1
24
PDB header: ribosomeChain: M: PDB Molecule: 40s ribosomal protein s18 (s13p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
17 d1bc9a_
8.7
18
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain18 c3dkqB_
8.7
41
PDB header: oxidoreductaseChain: B: PDB Molecule: pkhd-type hydroxylase sbal_3634;PDBTitle: crystal structure of putative oxygenase (yp_001051978.1) from2 shewanella baltica os155 at 2.26 a resolution
19 d1zira2
8.6
32
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins20 c3knyA_
8.5
20
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein bt_3535;PDBTitle: crystal structure of a two domain protein with unknown function2 (bt_3535) from bacteroides thetaiotaomicron vpi-5482 at 2.60 a3 resolution
21 c2zkrq_
not modelled
8.5
50
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: rna expansion segment es31 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
22 d1elpa1
not modelled
8.3
37
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins23 d2b50a1
not modelled
8.2
14
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain24 c2ovqA_
not modelled
8.2
32
PDB header: transcription/cell cycleChain: A: PDB Molecule: s-phase kinase-associated protein 1a;PDBTitle: structure of the skp1-fbw7-cyclinedegc complex
25 c1s1iQ_
not modelled
7.9
60
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l21-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
26 d2c9wc1
not modelled
7.8
24
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain27 c3drzE_
not modelled
7.3
25
PDB header: unknown functionChain: E: PDB Molecule: btb/poz domain-containing protein kctd5;PDBTitle: x-ray crystal structure of the n-terminal btb domain of human kctd52 protein
28 c2hdeA_
not modelled
7.1
17
PDB header: transcriptionChain: A: PDB Molecule: histone deacetylase complex subunit sap18;PDBTitle: solution structure of human sap18
29 c2vx6A_
not modelled
6.9
16
PDB header: hydrolaseChain: A: PDB Molecule: cellvibrio japonicus mannanase cjman26c;PDBTitle: cellvibrio japonicus mannanase cjman26c gal1man4-bound form
30 c2dbiA_
not modelled
6.5
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ybiu;PDBTitle: crystal structure of a hypothetical protein jw0805 from2 escherichia coli
31 c3g36D_
not modelled
6.4
10
PDB header: nuclear proteinChain: D: PDB Molecule: protein dpy-30 homolog;PDBTitle: crystal structure of the human dpy-30-like c-terminal domain
32 d1qcsa1
not modelled
6.3
36
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like33 c1wd6B_
not modelled
6.3
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein ydhr;PDBTitle: crystal structure of jw1657 from escherichia coli
34 d2eyqa1
not modelled
6.2
22
Fold: SH3-like barrelSuperfamily: CarD-likeFamily: CarD-like35 d2csga1
not modelled
6.0
27
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: YbiU-like36 c4a1aP_
not modelled
5.9
50
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l21;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
37 c1okiB_
not modelled
5.8
26
PDB header: eye lens proteinChain: B: PDB Molecule: beta crystallin b1;PDBTitle: crystal structure of truncated human beta-b1-crystallin
38 c2ddxA_
not modelled
5.7
22
PDB header: hydrolaseChain: A: PDB Molecule: beta-1,3-xylanase;PDBTitle: crystal structure of beta-1,3-xylanase from vibrio sp. ax-4
39 c3gf8A_
not modelled
5.7
36
PDB header: carbohydrate binding proteinChain: A: PDB Molecule: putative polysaccharide binding proteins (duf1812);PDBTitle: crystal structure of putative polysaccharide binding proteins2 (duf1812) (np_809975.1) from bacteroides thetaiotaomicron vpi-5482 at3 2.20 a resolution
40 c2bv2B_
not modelled
5.7
16
PDB header: crystallinChain: B: PDB Molecule: ciona betagamma-crystallin;PDBTitle: beta gamma crystallin from ciona intestinalis
41 d1nhya2
not modelled
5.7
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain42 d1vj1a1
not modelled
5.5
17
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain43 c3ogiC_
not modelled
5.5
100
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative esat-6-like protein 6;PDBTitle: crystal structure of the mycobacterium tuberculosis h37rv esxop2 complex (rv2346c-rv2347c)
44 d1okia1
not modelled
5.5
26
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins45 c3izcU_
not modelled
5.4
60
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein rpl21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
46 d1pbva_
not modelled
5.3
12
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain47 c2zkqm_
not modelled
5.3
23
PDB header: ribosomal protein/rnaChain: M: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
48 d2hiqa1
not modelled
5.2
25
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Hypothetical protein YdhR49 d1pvda3
not modelled
5.1
24
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module50 c2oaxC_
not modelled
5.1
33
PDB header: transcriptionChain: C: PDB Molecule: mineralocorticoid receptor;PDBTitle: crystal structure of the s810l mutant mineralocorticoid2 receptor associated with sc9420