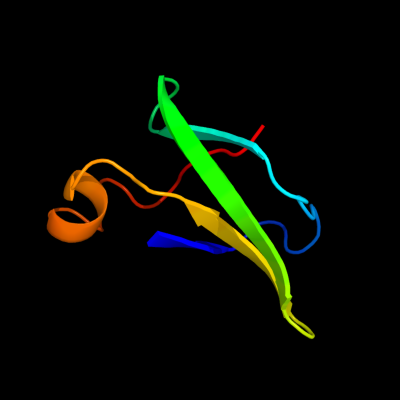
| 1 | d1pj5a1
|
|
 |
93.3 |
16 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
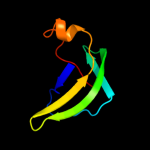
| 2 | d1v5va1
|
|
 |
91.2 |
22 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
| 3 | c2j5uB_
|
|
 |
90.8 |
20 |
PDB header:cell shape regulation
Chain: B: PDB Molecule:mrec protein;
PDBTitle: mrec lysteria monocytogenes
|
| 4 | d1wosa1
|
|
 |
88.2 |
15 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
| 5 | c1v5vA_
|
|
 |
88.1 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of a component of glycine cleavage system: t-protein2 from pyrococcus horikoshii ot3 at 1.5 a resolution
|
| 6 | d1vloa1
|
|
 |
78.9 |
20 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
| 7 | c1yx2B_
|
|
 |
76.7 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of the probable aminomethyltransferase2 from bacillus subtilis
|
| 8 | c1worA_
|
|
 |
70.4 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of t-protein of the glycine cleavage2 system
|
| 9 | c2qf4A_
|
|
 |
65.5 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:cell shape determining protein mrec;
PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
|
| 10 | c3girA_
|
|
 |
60.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of glycine cleavage system2 aminomethyltransferase t from bartonella henselae
|
| 11 | c3tfhB_
|
|
 |
59.5 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:gcvt-like aminomethyltransferase protein;
PDBTitle: dmsp-dependent demethylase from p. ubique - apo
|
| 12 | c1wsrA_
|
|
 |
58.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of human t-protein of glycine cleavage2 system
|
| 13 | d1c99a_
|
|
 |
47.9 |
17 |
Fold:Transmembrane helix hairpin
Superfamily:F1F0 ATP synthase subunit C
Family:F1F0 ATP synthase subunit C |
| 14 | c1pj6A_
|
|
 |
46.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n,n-dimethylglycine oxidase;
PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
|
| 15 | d1fftb2
|
|
 |
36.9 |
15 |
Fold:Transmembrane helix hairpin
Superfamily:Cytochrome c oxidase subunit II-like, transmembrane region
Family:Cytochrome c oxidase subunit II-like, transmembrane region |
| 16 | c1txkA_
|
|
 |
35.2 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:glucans biosynthesis protein g;
PDBTitle: crystal structure of escherichia coli opgg
|
| 17 | d1jz8a3
|
|
 |
33.6 |
17 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:beta-Galactosidase/glucuronidase, N-terminal domain |
| 18 | d1txka2
|
|
 |
33.2 |
23 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:MdoG-like |
| 19 | c1vloA_
|
|
 |
32.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of aminomethyltransferase (t protein;2 tetrahydrofolate-dependent) of glycine cleavage system (np417381)3 from escherichia coli k12 at 1.70 a resolution
|
| 20 | c1yo8A_
|
|
 |
27.1 |
13 |
PDB header:cell adhesion
Chain: A: PDB Molecule:thrombospondin-2;
PDBTitle: structure of the c-terminal domain of human thrombospondin-2
|
| 21 | c3o0lB_ |
|
not modelled |
26.7 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf1425 family member (shew_1734) from2 shewanella sp. pv-4 at 1.81 a resolution
|
| 22 | d1yq2a3 |
|
not modelled |
26.1 |
13 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:beta-Galactosidase/glucuronidase, N-terminal domain |
| 23 | c1kqsA_ |
|
not modelled |
24.8 |
23 |
PDB header:ribosome
Chain: A: PDB Molecule:ribosomal protein l2;
PDBTitle: the haloarcula marismortui 50s complexed with a2 pretranslocational intermediate in protein synthesis
|
| 24 | c3cb0B_ |
|
not modelled |
24.5 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxyphenylacetate 3-monooxygenase;
PDBTitle: cobr
|
| 25 | c1vlyA_ |
|
not modelled |
24.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:unknown protein from 2d-page;
PDBTitle: crystal structure of a putative aminomethyltransferase (ygfz) from2 escherichia coli at 1.30 a resolution
|
| 26 | d1wgba_ |
|
not modelled |
22.2 |
15 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 27 | d1kkga_ |
|
not modelled |
19.9 |
38 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Ribosome-binding factor A, RbfA
Family:Ribosome-binding factor A, RbfA |
| 28 | c2c1lA_ |
|
not modelled |
18.8 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: structure of the bfii restriction endonuclease
|
| 29 | d1pa4a_ |
|
not modelled |
18.2 |
44 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Ribosome-binding factor A, RbfA
Family:Ribosome-binding factor A, RbfA |
| 30 | c2kzfA_ |
|
not modelled |
18.1 |
38 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:ribosome-binding factor a;
PDBTitle: solution nmr structure of the thermotoga maritima protein tm0855 a2 putative ribosome binding factor a
|
| 31 | c2r6vA_ |
|
not modelled |
18.0 |
15 |
PDB header:flavoprotein
Chain: A: PDB Molecule:uncharacterized protein ph0856;
PDBTitle: crystal structure of fmn-binding protein (np_142786.1) from pyrococcus2 horikoshii at 1.35 a resolution
|
| 32 | c2r39A_ |
|
not modelled |
17.2 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:fixg-related protein;
PDBTitle: crystal structure of fixg-related protein from vibrio parahaemolyticus
|
| 33 | c2r0xA_ |
|
not modelled |
16.9 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:possible flavin reductase;
PDBTitle: crystal structure of a putative flavin reductase (ycdh, hs_1225) from2 haemophilus somnus 129pt at 1.06 a resolution
|
| 34 | d2dyja1 |
|
not modelled |
16.6 |
34 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Ribosome-binding factor A, RbfA
Family:Ribosome-binding factor A, RbfA |
| 35 | d1bhua_ |
|
not modelled |
15.8 |
20 |
Fold:gamma-Crystallin-like
Superfamily:gamma-Crystallin-like
Family:Streptomyces metalloproteinase inhibitor, SMPI |
| 36 | d1v58a2 |
|
not modelled |
15.2 |
21 |
Fold:Cystatin-like
Superfamily:DsbC/DsbG N-terminal domain-like
Family:DsbC/DsbG N-terminal domain-like |
| 37 | c3fljA_ |
|
not modelled |
14.8 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein conserved in bacteria with a
PDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 with a cystatin-like fold (yp_168589.1) from silicibacter pomeroyi3 dss-3 at 2.00 a resolution
|
| 38 | c1w2tE_ |
|
not modelled |
14.4 |
10 |
PDB header:hydrolase
Chain: E: PDB Molecule:beta fructosidase;
PDBTitle: beta-fructosidase from thermotoga maritima in complex with2 raffinose
|
| 39 | c2kebA_ |
|
not modelled |
14.4 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna polymerase subunit alpha b;
PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
|
| 40 | c2zjbB_ |
|
not modelled |
13.3 |
35 |
PDB header:recombination
Chain: B: PDB Molecule:meiotic recombination protein dmc1/lim15 homolog;
PDBTitle: crystal structure of the human dmc1-m200v polymorphic2 variant
|
| 41 | d1josa_ |
|
not modelled |
12.4 |
35 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Ribosome-binding factor A, RbfA
Family:Ribosome-binding factor A, RbfA |
| 42 | c3o5aB_ |
|
not modelled |
12.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:diheme cytochrome c napb;
PDBTitle: crystal structure of partially reduced periplasmic nitrate reductase2 from cupriavidus necator using ionic liquids
|
| 43 | c2p0yA_ |
|
not modelled |
12.1 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr6
|
| 44 | c2d38A_ |
|
not modelled |
11.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical nadh-dependent fmn oxidoreductase;
PDBTitle: the crystal structure of flavin reductase hpac complexed with nadp+
|
| 45 | d1whva_ |
|
not modelled |
11.6 |
30 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 46 | d1v5wa_ |
|
not modelled |
11.1 |
35 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 47 | c1pznG_ |
|
not modelled |
10.9 |
43 |
PDB header:recombination
Chain: G: PDB Molecule:dna repair and recombination protein rad51;
PDBTitle: rad51 (rada)
|
| 48 | d1pzna2 |
|
not modelled |
10.6 |
43 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 49 | d2e7ga1 |
|
not modelled |
10.6 |
19 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Ribosome-binding factor A, RbfA
Family:Ribosome-binding factor A, RbfA |
| 50 | c2w0mA_ |
|
not modelled |
10.5 |
43 |
PDB header:unknown function
Chain: A: PDB Molecule:sso2452;
PDBTitle: crystal structure of sso2452 from sulfolobus solfataricus2 p2
|
| 51 | d1rz0a_ |
|
not modelled |
10.4 |
18 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 52 | c3nmbA_ |
|
not modelled |
10.3 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sugar hydrolase;
PDBTitle: crystal structure of a putative sugar hydrolase (bacova_03189) from2 bacteroides ovatus at 2.40 a resolution
|
| 53 | d2axte1 |
|
not modelled |
9.8 |
30 |
Fold:Single transmembrane helix
Superfamily:Cytochrome b559 subunits
Family:Cytochrome b559 subunits |
| 54 | c3hbkA_ |
|
not modelled |
9.7 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative glycosyl hydrolase;
PDBTitle: crystal structure of putative glycosyl hydrolase, was domain of2 unknown function (duf1080) (yp_001302580.1) from parabacteroides3 distasonis atcc 8503 at 2.36 a resolution
|
| 55 | c3uaiA_ |
|
not modelled |
9.7 |
18 |
PDB header:isomerase/chaperone
Chain: A: PDB Molecule:h/aca ribonucleoprotein complex subunit 4;
PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
|
| 56 | c3izcN_ |
|
not modelled |
9.6 |
22 |
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein rpl14 (l14e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 57 | c2jqoA_ |
|
not modelled |
9.6 |
13 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical protein yoba;
PDBTitle: nmr solution structure of bacillus subtilis yoba 21-120:2 northeast structural genomics consortium target sr547
|
| 58 | c1pkvB_ |
|
not modelled |
9.4 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
| 59 | c1pkvA_ |
|
not modelled |
9.4 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
| 60 | c3h3lB_ |
|
not modelled |
9.2 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative sugar hydrolase;
PDBTitle: crystal structure of putative sugar hydrolase (yp_001304206.1) from2 parabacteroides distasonis atcc 8503 at 1.59 a resolution
|
| 61 | d1mvqa_ |
|
not modelled |
9.1 |
23 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Legume lectins |
| 62 | d2evra1 |
|
not modelled |
8.6 |
25 |
Fold:SH3-like barrel
Superfamily:Prokaryotic SH3-related domain
Family:Spr N-terminal domain-like |
| 63 | d1i8da1 |
|
not modelled |
8.6 |
22 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
| 64 | d2cp6a1 |
|
not modelled |
8.5 |
15 |
Fold:SH3-like barrel
Superfamily:Cap-Gly domain
Family:Cap-Gly domain |
| 65 | d2i00a1 |
|
not modelled |
8.5 |
11 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
| 66 | d1h9wa_ |
|
not modelled |
8.4 |
20 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Legume lectins |
| 67 | d2i1qa2 |
|
not modelled |
8.3 |
39 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 68 | d2cu6a1 |
|
not modelled |
8.3 |
14 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:PaaD-like |
| 69 | d1zy9a1 |
|
not modelled |
8.3 |
43 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:YicI N-terminal domain-like |
| 70 | d1zq1a1 |
|
not modelled |
8.3 |
21 |
Fold:Sm-like fold
Superfamily:GatD N-terminal domain-like
Family:GatD N-terminal domain-like |
| 71 | d2hv2a1 |
|
not modelled |
8.2 |
12 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
| 72 | c1icfB_ |
|
not modelled |
8.1 |
38 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein (cathepsin l: light chain);
PDBTitle: crystal structure of mhc class ii associated p41 ii fragment in2 complex with cathepsin l
|
| 73 | d2pmaa1 |
|
not modelled |
8.0 |
16 |
Fold:Acid proteases
Superfamily:Acid proteases
Family:LPG0085-like |
| 74 | d2cb2a1 |
|
not modelled |
7.9 |
25 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:SOR-like |
| 75 | c3k87B_ |
|
not modelled |
7.9 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:chlorophenol-4-monooxygenase component 1;
PDBTitle: crystal structure of nadh:fad oxidoreductase (tftc) - fad2 complex
|
| 76 | c3u1xA_ |
|
not modelled |
7.8 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative glycosyl hydrolase;
PDBTitle: crystal structure of a putative glycosyl hydrolase (bdi_1869) from2 parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
| 77 | c3khsB_ |
|
not modelled |
7.8 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 78 | d1vm8a_ |
|
not modelled |
7.6 |
26 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 79 | c2z8nB_ |
|
not modelled |
7.6 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:27.5 kda virulence protein;
PDBTitle: structural basis for the catalytic mechanism of phosphothreonine lyase
|
| 80 | d2cnaa_ |
|
not modelled |
7.5 |
22 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Legume lectins |
| 81 | d1mzaa_ |
|
not modelled |
7.5 |
22 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 82 | d1tf7a2 |
|
not modelled |
7.5 |
43 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 83 | c2l66B_ |
|
not modelled |
7.4 |
17 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, abrb family;
PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
|
| 84 | c3nfgG_ |
|
not modelled |
7.4 |
10 |
PDB header:transcription
Chain: G: PDB Molecule:dna-directed rna polymerase i subunit rpa49;
PDBTitle: crystal structure of dimerization module of rna polymerase i2 subcomplex a49/a34.5
|
| 85 | d1g7sa1 |
|
not modelled |
7.3 |
16 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Elongation factors |
| 86 | d1o7ia_ |
|
not modelled |
7.3 |
23 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 87 | c1x31A_ |
|
not modelled |
7.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sarcosine oxidase alpha subunit;
PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
|
| 88 | c2v31A_ |
|
not modelled |
7.3 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-activating enzyme e1 x;
PDBTitle: structure of first catalytic cysteine half-domain of mouse2 ubiquitin-activating enzyme
|
| 89 | d1eu3a1 |
|
not modelled |
7.2 |
19 |
Fold:OB-fold
Superfamily:Bacterial enterotoxins
Family:Superantigen toxins, N-terminal domain |
| 90 | d2uubq1 |
|
not modelled |
7.2 |
7 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 91 | c2xlfA_ |
|
not modelled |
7.2 |
12 |
PDB header:metal binding protein
Chain: A: PDB Molecule:sll1785 protein;
PDBTitle: structure and metal-loading of a soluble periplasm cupro-protein:2 apo-cuca-closed (semet)
|
| 92 | c2eqnA_ |
|
not modelled |
7.0 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:hypothetical protein loc92345;
PDBTitle: solution structure of the naf1 domain of hypothetical2 protein bc008207 [homo sapiens]
|
| 93 | c4a19F_ |
|
not modelled |
7.0 |
19 |
PDB header:ribosome
Chain: F: PDB Molecule:rpl14;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
|
| 94 | c2kncA_ |
|
not modelled |
6.9 |
19 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 95 | c1jniA_ |
|
not modelled |
6.8 |
0 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diheme cytochrome c napb;
PDBTitle: structure of the napb subunit of the periplasmic nitrate2 reductase from haemophilus influenzae.
|
| 96 | d1jnia_ |
|
not modelled |
6.8 |
0 |
Fold:Multiheme cytochromes
Superfamily:Multiheme cytochromes
Family:Di-heme elbow motif |
| 97 | c1cn3F_ |
|
not modelled |
6.8 |
43 |
PDB header:viral protein
Chain: F: PDB Molecule:fragment of coat protein vp2;
PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
|
| 98 | c2ztsB_ |
|
not modelled |
6.7 |
43 |
PDB header:atp-binding protein
Chain: B: PDB Molecule:putative uncharacterized protein ph0186;
PDBTitle: crystal structure of kaic-like protein ph0186 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
|
| 99 | d1iaua_ |
|
not modelled |
6.7 |
14 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |