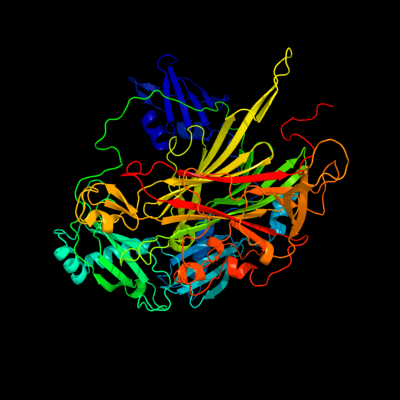
| 1 | c1d6uB_
|
|
 |
100.0 |
100 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure of e. coli amine oxidase anaerobically reduced with2 beta-phenylethylamine
|
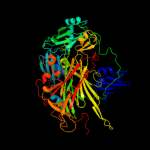
| 2 | c1ksiA_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure of a eukaryotic (pea seedling) copper-containing2 amine oxidase at 2.2a resolution
|
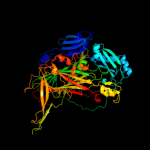
| 3 | c1ekmC_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure at 2.5 a resolution of zinc-substituted2 copper amine oxidase of hansenula polymorpha expressed in3 escherichia coli
|
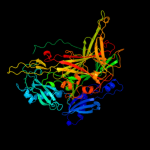
| 4 | c1ui7A_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phenylethylamine oxidase;
PDBTitle: site-directed mutagenesis of his433 involved in binding of2 copper ion in arthrobacter globiformis amine oxidase
|
| 5 | c3nbbC_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:peroxisomal primary amine oxidase;
PDBTitle: crystal structure of mutant y305f expressed in e. coli in the copper2 amine oxidase from hansenula polymorpha
|
| 6 | c3loyB_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:copper amine oxidase;
PDBTitle: crystal structure of a copper-containing benzylamine oxidase from2 hansenula polymorpha
|
| 7 | c3higB_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amiloride-sensitive amine oxidase;
PDBTitle: crystal structure of human diamine oxidase in complex with the2 inhibitor berenil
|
| 8 | c3pgbA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of aspergillus nidulans amine oxidase
|
| 9 | c1n9eA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysyl oxidase;
PDBTitle: crystal structure of pichia pastoris lysyl oxidase pplo
|
| 10 | c1w7cA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysyl oxidase;
PDBTitle: pplo at 1.23 angstroms
|
| 11 | c2pncB_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:copper amine oxidase, liver isozyme;
PDBTitle: crystal structure of bovine plasma copper-containing amine oxidase in2 complex with clonidine
|
| 12 | d1d6za1
|
|
 |
100.0 |
100 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 13 | d1w6ga1
|
|
 |
100.0 |
31 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 14 | d2oqea1
|
|
 |
100.0 |
29 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 15 | d1w2za1
|
|
 |
100.0 |
32 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 16 | d1w7ca1
|
|
 |
100.0 |
25 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 17 | d1d6za3
|
|
 |
100.0 |
100 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 18 | d1w6ga3
|
|
 |
100.0 |
29 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 19 | d1w2za3
|
|
 |
100.0 |
20 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 20 | d1d6za2
|
|
 |
99.9 |
100 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 21 | d2oqea2 |
|
not modelled |
99.9 |
26 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 22 | d2oqea3 |
|
not modelled |
99.9 |
23 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 23 | d1w2za2 |
|
not modelled |
99.9 |
24 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 24 | d1w6ga2 |
|
not modelled |
99.8 |
23 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 25 | d1w7ca2 |
|
not modelled |
99.4 |
19 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 26 | d1d6za4 |
|
not modelled |
98.7 |
100 |
Fold:N domain of copper amine oxidase-like
Superfamily:Copper amine oxidase, domain N
Family:Copper amine oxidase, domain N |
| 27 | c3mlhA_ |
|
not modelled |
40.0 |
21 |
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: crystal structure of the 2009 h1n1 influenza virus hemagglutinin2 receptor-binding domain
|
| 28 | d2viua_ |
|
not modelled |
38.5 |
24 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 29 | d1kx5d_ |
|
not modelled |
36.4 |
24 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 30 | d1eqzb_ |
|
not modelled |
34.7 |
22 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 31 | c3qqiB_ |
|
not modelled |
32.0 |
30 |
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin;
PDBTitle: crystal structure of the ha1 receptor binding domain of h22 hemagglutinin
|
| 32 | d1rvxa_ |
|
not modelled |
30.7 |
28 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 33 | d1s32d_ |
|
not modelled |
30.0 |
24 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 34 | c3imoC_ |
|
not modelled |
29.7 |
26 |
PDB header:unknown function
Chain: C: PDB Molecule:integron cassette protein;
PDBTitle: structure from the mobile metagenome of vibrio cholerae.2 integron cassette protein vch_cass14
|
| 35 | d1rv0h_ |
|
not modelled |
28.5 |
21 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 36 | d1w7ca3 |
|
not modelled |
28.1 |
16 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 37 | c1ha0A_ |
|
not modelled |
27.9 |
24 |
PDB header:viral protein
Chain: A: PDB Molecule:protein (hemagglutinin precursor);
PDBTitle: hemagglutinin precursor ha0
|
| 38 | d2dira1 |
|
not modelled |
26.7 |
22 |
Fold:THUMP domain
Superfamily:THUMP domain-like
Family:Minimal THUMP |
| 39 | c1x9yD_ |
|
not modelled |
23.3 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:cysteine proteinase;
PDBTitle: the prostaphopain b structure
|
| 40 | d1uwwa_ |
|
not modelled |
21.9 |
17 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Family 28 carbohydrate binding module, CBM28 |
| 41 | d1mqma_ |
|
not modelled |
21.2 |
24 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 42 | d1jsda_ |
|
not modelled |
21.0 |
23 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 43 | d2cx1a2 |
|
not modelled |
20.3 |
20 |
Fold:Cystatin-like
Superfamily:Pre-PUA domain
Family:Hypothetical protein APE0525, N-terminal domain |
| 44 | c2jssA_ |
|
not modelled |
20.1 |
23 |
PDB header:chaperone/nuclear protein
Chain: A: PDB Molecule:chimera of histone h2b.1 and histone h2a.z;
PDBTitle: nmr structure of chaperone chz1 complexed with histone2 h2a.z-h2b
|
| 45 | d1rd8a_ |
|
not modelled |
20.0 |
23 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 46 | c2cfoA_ |
|
not modelled |
19.5 |
35 |
PDB header:ligase
Chain: A: PDB Molecule:glutamyl-trna synthetase;
PDBTitle: non-discriminating glutamyl-trna synthetase from2 thermosynechococcus elongatus in complex with glu
|
| 47 | d2plga1 |
|
not modelled |
17.6 |
39 |
Fold:Secretion chaperone-like
Superfamily:Type III secretory system chaperone-like
Family:Tll0839-like |
| 48 | c3al0C_ |
|
not modelled |
16.8 |
27 |
PDB header:ligase/rna
Chain: C: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit c, glutamyl-
PDBTitle: crystal structure of the glutamine transamidosome from thermotoga2 maritima in the glutamylation state.
|
| 49 | d2visc_ |
|
not modelled |
16.0 |
23 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 50 | d1nkqa_ |
|
not modelled |
16.0 |
40 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 51 | d1jsma_ |
|
not modelled |
12.9 |
30 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 52 | c2rfuA_ |
|
not modelled |
12.8 |
24 |
PDB header:viral protein
Chain: A: PDB Molecule:influenza b hemagglutinin (ha);
PDBTitle: crystal structure of influenza b virus hemagglutinin in complex with2 lstc receptor analog
|
| 53 | c3rurB_ |
|
not modelled |
12.8 |
19 |
PDB header:metal transport
Chain: B: PDB Molecule:iron-regulated surface determinant protein b;
PDBTitle: staphylococcus aureus heme-bound selenomethionine-labeled isdb-n2
|
| 54 | d1jp4a_ |
|
not modelled |
12.6 |
31 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 55 | c1zxuA_ |
|
not modelled |
12.1 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:at5g01750 protein;
PDBTitle: x-ray structure of protein from arabidopsis thaliana2 at5g01750
|
| 56 | d2q4ma1 |
|
not modelled |
12.1 |
7 |
Fold:Tubby C-terminal domain-like
Superfamily:Tubby C-terminal domain-like
Family:At5g01750-like |
| 57 | d1pfsa_ |
|
not modelled |
10.2 |
50 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Phage ssDNA-binding proteins |
| 58 | c2ka6B_ |
|
not modelled |
9.8 |
19 |
PDB header:transcription regulator
Chain: B: PDB Molecule:signal transducer and activator of transcription
PDBTitle: nmr structure of the cbp-taz2/stat1-tad complex
|
| 59 | d1r5ya_ |
|
not modelled |
9.6 |
4 |
Fold:TIM beta/alpha-barrel
Superfamily:tRNA-guanine transglycosylase
Family:tRNA-guanine transglycosylase |
| 60 | c3k1tA_ |
|
not modelled |
9.5 |
55 |
PDB header:ligase
Chain: A: PDB Molecule:glutamate--cysteine ligase gsha;
PDBTitle: crystal structure of putative gamma-glutamylcysteine synthetase2 (yp_546622.1) from methylobacillus flagellatus kt at 1.90 a3 resolution
|
| 61 | c2ja2A_ |
|
not modelled |
9.4 |
33 |
PDB header:ligase
Chain: A: PDB Molecule:glutamyl-trna synthetase;
PDBTitle: mycobacterium tuberculosis glutamyl-trna synthetase
|
| 62 | c3acgA_ |
|
not modelled |
9.3 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-1,4-endoglucanase;
PDBTitle: crystal structure of carbohydrate-binding module family 282 from clostridium josui cel5a in complex with cellobiose
|
| 63 | d2d9ra1 |
|
not modelled |
9.3 |
8 |
Fold:Double-split beta-barrel
Superfamily:AF2212/PG0164-like
Family:PG0164-like |
| 64 | d1xxxa1 |
|
not modelled |
9.2 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 65 | c3bbnI_ |
|
not modelled |
9.2 |
26 |
PDB header:ribosome
Chain: I: PDB Molecule:ribosomal protein s9;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 66 | d2vqei1 |
|
not modelled |
9.0 |
42 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 67 | d2cs7a1 |
|
not modelled |
8.9 |
35 |
Fold:IL8-like
Superfamily:PhtA domain-like
Family:PhtA domain-like |
| 68 | d2ibxa1 |
|
not modelled |
8.8 |
29 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Influenza hemagglutinin headpiece |
| 69 | c2xfyA_ |
|
not modelled |
8.8 |
52 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-amylase;
PDBTitle: crystal structure of barley beta-amylase complexed with2 alpha-cyclodextrin
|
| 70 | d1hcza1 |
|
not modelled |
8.7 |
22 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Cytochrome f, large domain
Family:Cytochrome f, large domain |
| 71 | c3cu4A_ |
|
not modelled |
8.6 |
16 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome c family protein;
PDBTitle: omcf, outer membrance cytochrome f from geobacter2 sulfurreducens
|
| 72 | d2o8ra4 |
|
not modelled |
8.6 |
7 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 73 | c1fllA_ |
|
not modelled |
8.5 |
17 |
PDB header:apoptosis
Chain: A: PDB Molecule:tnf receptor associated factor 3;
PDBTitle: molecular basis for cd40 signaling mediated by traf3
|
| 74 | d1jjcb4 |
|
not modelled |
8.4 |
4 |
Fold:Ferredoxin-like
Superfamily:Anticodon-binding domain of PheRS
Family:Anticodon-binding domain of PheRS |
| 75 | d2dy1a4 |
|
not modelled |
8.4 |
26 |
Fold:Ferredoxin-like
Superfamily:EF-G C-terminal domain-like
Family:EF-G/eEF-2 domains III and V |
| 76 | c3gr0D_ |
|
not modelled |
8.2 |
14 |
PDB header:membrane protein
Chain: D: PDB Molecule:protein prgh;
PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
|
| 77 | c1zljE_ |
|
not modelled |
8.1 |
28 |
PDB header:transcription
Chain: E: PDB Molecule:dormancy survival regulator;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
|
| 78 | d1ejxb_ |
|
not modelled |
8.1 |
27 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 79 | c2yxgD_ |
|
not modelled |
8.1 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 80 | c1g59A_ |
|
not modelled |
8.1 |
45 |
PDB header:ligase/rna
Chain: A: PDB Molecule:glutamyl-trna synthetase;
PDBTitle: glutamyl-trna synthetase complexed with trna(glu).
|
| 81 | c3afhA_ |
|
not modelled |
8.0 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:glutamyl-trna synthetase 2;
PDBTitle: crystal structure of thermotoga maritima nondiscriminating glutamyl-2 trna synthetase in complex with a glutamyl-amp analog
|
| 82 | c3iz6I_ |
|
not modelled |
7.8 |
42 |
PDB header:ribosome
Chain: I: PDB Molecule:40s ribosomal protein s16 (s9p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 83 | c3sbrF_ |
|
not modelled |
7.6 |
8 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:nitrous-oxide reductase;
PDBTitle: pseudomonas stutzeri nitrous oxide reductase, p1 crystal form with2 substrate
|
| 84 | c2xzmI_ |
|
not modelled |
7.6 |
42 |
PDB header:ribosome
Chain: I: PDB Molecule:rps16e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 85 | d2oo3a1 |
|
not modelled |
7.6 |
7 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:LPG1296-like |
| 86 | d2gy9i1 |
|
not modelled |
7.5 |
37 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 87 | c3si9B_ |
|
not modelled |
7.5 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 88 | c3luzA_ |
|
not modelled |
7.5 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:extragenic suppressor protein suhb;
PDBTitle: crystal structure of extragenic suppressor protein suhb from2 bartonella henselae, via combined iodide sad molecular replacement
|
| 89 | d1owxa_ |
|
not modelled |
7.5 |
17 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 90 | c1y4hA_ |
|
not modelled |
7.4 |
12 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:cysteine protease;
PDBTitle: wild type staphopain-staphostatin complex
|
| 91 | c2ehhE_ |
|
not modelled |
7.4 |
20 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 92 | c3pueA_ |
|
not modelled |
7.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 93 | d1a6ca1 |
|
not modelled |
7.3 |
29 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Comoviridae-like VP |
| 94 | c1kzzA_ |
|
not modelled |
7.3 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:tnf receptor associated factor 3;
PDBTitle: downstream regulator tank binds to the cd40 recognition2 site on traf3
|
| 95 | c2vc6A_ |
|
not modelled |
7.3 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 96 | c3fluD_ |
|
not modelled |
7.2 |
32 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 97 | d1xi6a_ |
|
not modelled |
6.9 |
38 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 98 | d1lbva_ |
|
not modelled |
6.9 |
38 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 99 | d1w0pa2 |
|
not modelled |
6.9 |
12 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Vibrio cholerae sialidase, N-terminal and insertion domains |