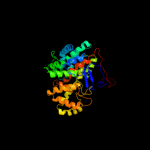
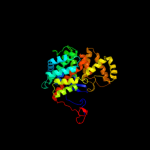
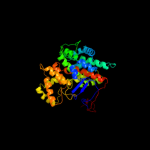
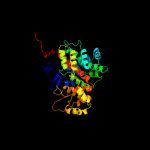
1 c2h12C_
100.0
31
PDB header: transferaseChain: C: PDB Molecule: citrate synthase;PDBTitle: structure of acetobacter aceti citrate synthase complexed2 with oxaloacetate and carboxymethyldethia coenzyme a (cmx)
2 d1k3pa_
100.0
30
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase3 d1ioma_
100.0
36
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase4 c2ibpB_
100.0
34
PDB header: transferaseChain: B: PDB Molecule: citrate synthase;PDBTitle: crystal structure of citrate synthase from pyrobaculum aerophilum
5 d1aj8a_
100.0
36
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase6 c3o8jH_
100.0
97
PDB header: transferaseChain: H: PDB Molecule: 2-methylcitrate synthase;PDBTitle: crystal structure of 2-methylcitrate synthase (prpc) from salmonella2 typhimurium
7 c3msuA_
100.0
30
PDB header: transferaseChain: A: PDB Molecule: citrate synthase;PDBTitle: crystal structure of citrate synthase from francisella tularensis
8 d1a59a_
100.0
41
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase9 c3hwkE_
100.0
38
PDB header: transferaseChain: E: PDB Molecule: methylcitrate synthase;PDBTitle: crystal structure of methylcitrate synthase from2 mycobacterium tuberculosis
10 c3tqgA_
100.0
52
PDB header: transferaseChain: A: PDB Molecule: 2-methylcitrate synthase;PDBTitle: structure of the 2-methylcitrate synthase (prpc) from coxiella2 burnetii
11 c1vgpA_
100.0
37
PDB header: transferaseChain: A: PDB Molecule: 373aa long hypothetical citrate synthase;PDBTitle: crystal structure of an isozyme of citrate synthase from sulfolbus2 tokodaii strain7
12 c2r26C_
100.0
35
PDB header: transferaseChain: C: PDB Molecule: citrate synthase;PDBTitle: the structure of the ternary complex of carboxymethyl2 coenzyme a and oxalateacetate with citrate synthase from3 the thermophilic archaeonthermoplasma acidophilum
13 c1vgmB_
100.0
34
PDB header: transferaseChain: B: PDB Molecule: 378aa long hypothetical citrate synthase;PDBTitle: crystal structure of an isozyme of citrate synthase from sulfolbus2 tokodaii strain7
14 c2p2wA_
100.0
38
PDB header: transferaseChain: A: PDB Molecule: citrate synthase;PDBTitle: crystal structure of citrate synthase from thermotoga maritima msb8
15 c2c6xA_
100.0
33
PDB header: transferaseChain: A: PDB Molecule: citrate synthase 1;PDBTitle: structure of bacillus subtilis citrate synthase
16 d1csha_
100.0
23
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase17 d2ctsa_
100.0
23
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase18 d1o7xa_
100.0
33
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase19 d1csca_
100.0
23
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase20 c2xzmS_
48.8
36
PDB header: ribosomeChain: S: PDB Molecule: rps15e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
21 d2huec1
not modelled
27.0
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones22 d1poib_
not modelled
24.8
12
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase beta subunit-like23 d1khda2
not modelled
22.2
21
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain24 d1kx5b_
not modelled
20.7
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones25 d1id3b_
not modelled
17.6
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones26 c2iouC_
not modelled
14.7
30
PDB header: viral protein/membrane proteinChain: C: PDB Molecule: major tropism determinant p1;PDBTitle: major tropism determinant p1 (mtd-p1) variant complexed with2 bordetella brochiseptica virulence factor pertactin extracellular3 domain (prn-e).
27 d1r3sa_
not modelled
12.3
15
Fold: TIM beta/alpha-barrelSuperfamily: UROD/MetE-likeFamily: Uroporphyrinogen decarboxylase, UROD28 c2kebA_
not modelled
11.9
18
PDB header: dna binding proteinChain: A: PDB Molecule: dna polymerase subunit alpha b;PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
29 d1yu0a2
not modelled
11.7
30
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Mtd variable domain30 d1keaa_
not modelled
11.0
16
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: Mismatch glycosylase31 d1hiod_
not modelled
10.6
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones32 d1orna_
not modelled
10.4
16
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: Endonuclease III33 c3ajvD_
not modelled
9.8
29
PDB header: hydrolaseChain: D: PDB Molecule: trna-splicing endonuclease;PDBTitle: splicing endonuclease from aeropyrum pernix
34 d1b8za_
not modelled
9.4
29
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein35 d1mula_
not modelled
8.3
23
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein36 c3c65A_
not modelled
8.0
20
PDB header: hydrolaseChain: A: PDB Molecule: uvrabc system protein c;PDBTitle: crystal structure of bacillus stearothermophilus uvrc 5'2 endonuclease domain
37 d2o97b1
not modelled
7.8
29
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein38 c1fctA_
not modelled
7.7
71
PDB header: transit peptideChain: A: PDB Molecule: ferredoxin chloroplastic transit peptidePDBTitle: nmr structures of ferredoxin chloroplastic transit peptide2 from chlamydomonas reinhardtii promoted by3 trifluoroethanol in aqueous solution
39 d1yu3a2
not modelled
7.7
30
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Mtd variable domain40 c2l5aA_
not modelled
7.7
15
PDB header: nuclear proteinChain: A: PDB Molecule: histone h3-like centromeric protein cse4, protein scm3,PDBTitle: structural basis for recognition of centromere specific histone h32 variant by nonhistone scm3
41 d2abka_
not modelled
7.1
12
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: Endonuclease III42 d1fhea1
not modelled
6.8
14
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain43 c2q6vA_
not modelled
6.5
13
PDB header: transferaseChain: A: PDB Molecule: glucuronosyltransferase gumk;PDBTitle: crystal structure of gumk in complex with udp
44 c1jpkA_
not modelled
6.5
11
PDB header: lyaseChain: A: PDB Molecule: uroporphyrinogen decarboxylase;PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
45 c3kgkA_
not modelled
6.2
46
PDB header: chaperoneChain: A: PDB Molecule: arsenical resistance operon trans-acting repressor arsd;PDBTitle: crystal structure of arsd
46 c3ktbD_
not modelled
6.1
15
PDB header: transcription regulatorChain: D: PDB Molecule: arsenical resistance operon trans-acting repressor;PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
47 c3ts3D_
not modelled
5.9
24
PDB header: viral proteinChain: D: PDB Molecule: capsid polyprotein;PDBTitle: crystal structure of the projection domain of the turkey astrovirus2 capsid protein at 1.5 angstrom resolution
48 d1o17a1
not modelled
5.8
18
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain49 d1brwa2
not modelled
5.7
13
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain50 d1m6ya1
not modelled
5.7
19
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain51 c1j9zB_
not modelled
5.7
22
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-cytochrome p450 reductase;PDBTitle: cypor-w677g
52 d1wg8a1
not modelled
5.6
17
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain53 c2nv4A_
not modelled
5.6
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0066 protein af_0241;PDBTitle: crystal structure of upf0066 protein af0241 in complex with2 s-adenosylmethionine. northeast structural genomics3 consortium target gr27
54 c2l0cA_
not modelled
5.4
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative membrane protein;PDBTitle: solution nmr structure of protein sty4237 (residues 36-120) from2 salmonella enterica, northeast structural genomics consortium target3 slr115
55 c2fheA_
not modelled
5.3
12
PDB header: transferase/substrateChain: A: PDB Molecule: glutathione s-transferase;PDBTitle: fasciola hepatica glutathione s-transferase isoform 1 in complex with2 glutathione
56 c3gndC_
not modelled
5.2
12
PDB header: lyaseChain: C: PDB Molecule: aldolase lsrf;PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
57 d2bv3a3
not modelled
5.1
20
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components58 c3ieyA_
not modelled
5.1
36
PDB header: hydrolase/rna binding proteinChain: A: PDB Molecule: trna-splicing endonuclease;PDBTitle: crystal structure of the functional nanoarchaeum equitans trna2 splicing endonuclease
59 d2elca2
not modelled
5.1
25
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain60 c2xzmV_
not modelled
5.0
24
PDB header: ribosomeChain: V: PDB Molecule: rps17e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1