1 c1miyB_
100.0
26
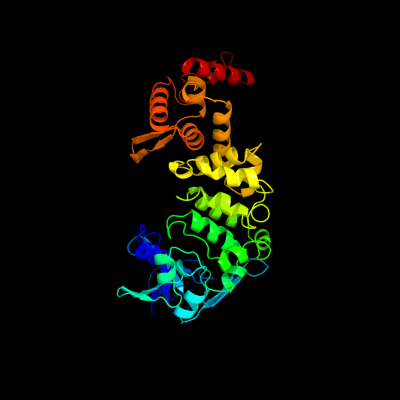
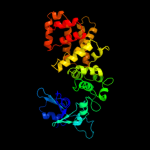
PDB header: translation, transferaseChain: B: PDB Molecule: trna cca-adding enzyme;PDBTitle: crystal structure of bacillus stearothermophilus cca-adding enzyme in2 complex with ctp
2 c1vfgB_
100.0
24
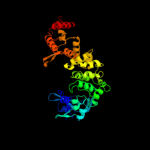
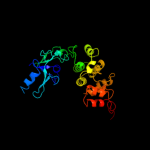
PDB header: transferase/rnaChain: B: PDB Molecule: poly a polymerase;PDBTitle: crystal structure of trna nucleotidyltransferase complexed2 with a primer trna and an incoming atp analog
3 c3h37B_
100.0
23
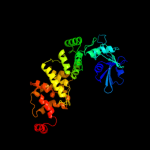
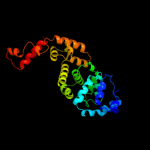
PDB header: transferaseChain: B: PDB Molecule: trna nucleotidyl transferase-related protein;PDBTitle: the structure of cca-adding enzyme apo form i
4 c3aqnA_
100.0
25
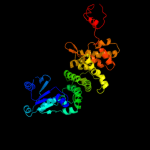
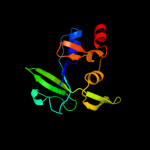
PDB header: transferaseChain: A: PDB Molecule: poly(a) polymerase;PDBTitle: complex structure of bacterial protein (apo form ii)
5 c1ou5A_
100.0
25
PDB header: translation, transferaseChain: A: PDB Molecule: trna cca-adding enzyme;PDBTitle: crystal structure of human cca-adding enzyme
6 d1miwa1
100.0
22
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like7 d1miwa2
100.0
36
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like8 d1vfga2
100.0
38
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like9 d1ou5a2
100.0
29
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like10 d1vfga1
100.0
18
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like11 d1ou5a1
99.9
23
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like12 d2qgsa1
97.3
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain13 d2pq7a1
97.2
17
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain14 d3djba1
96.6
22
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain15 d3b57a1
95.5
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain16 c3m5fA_
94.4
12
PDB header: hydrolaseChain: A: PDB Molecule: metal dependent phosphohydrolase;PDBTitle: structure of mj0384, a cas3 protein from methanocaldococcus jannaschii
17 d3dtoa1
94.3
18
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain18 c2la3A_
93.7
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr structure of the protein np_344798.1 reveals a cca-adding2 enzyme head domain
19 d1u6za1
91.8
16
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: Ppx associated domain20 d2pjqa1
91.7
14
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain21 d2fcla1
not modelled
91.4
19
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: TM1012-like22 c2q14A_
not modelled
90.9
19
PDB header: hydrolaseChain: A: PDB Molecule: phosphohydrolase;PDBTitle: crystal structure of phosphohydrolase (bt4208) from bacteroides2 thetaiotaomicron vpi-5482 at 2.20 a resolution
23 c2o08B_
not modelled
90.7
28
PDB header: hydrolaseChain: B: PDB Molecule: bh1327 protein;PDBTitle: crystal structure of a putative hd superfamily hydrolase (bh1327) from2 bacillus halodurans at 1.90 a resolution
24 c3ccgA_
not modelled
88.9
32
PDB header: hydrolaseChain: A: PDB Molecule: hd superfamily hydrolase;PDBTitle: crystal structure of predicted hd superfamily hydrolase involved in2 nad metabolism (np_347894.1) from clostridium acetobutylicum at 1.503 a resolution
25 c3skdA_
not modelled
87.7
18
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein tthb187;PDBTitle: crystal structure of the thermus thermophilus cas3 hd domain in the2 presence of ni2+
26 c3u1nC_
not modelled
86.6
25
PDB header: hydrolaseChain: C: PDB Molecule: sam domain and hd domain-containing protein 1;PDBTitle: structure of the catalytic core of human samhd1
27 c2ogiA_
not modelled
85.9
28
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein sag1661;PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
28 c3gw7A_
not modelled
85.5
14
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein yedj;PDBTitle: crystal structure of a metal-dependent phosphohydrolase2 with conserved hd domain (yedj) from escherichia coli in3 complex with nickel ions. northeast structural genomics4 consortium target er63
29 c2floA_
not modelled
84.9
15
PDB header: hydrolaseChain: A: PDB Molecule: exopolyphosphatase;PDBTitle: crystal structure of exopolyphosphatase (ppx) from e. coli o157:h7
30 d2o6ia1
not modelled
82.9
23
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain31 c2o6iA_
not modelled
82.9
23
PDB header: hydrolaseChain: A: PDB Molecule: hd domain protein;PDBTitle: structure of an enterococcus faecalis hd domain phosphohydrolase
32 d2gz4a1
not modelled
70.6
12
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain33 c3kq5A_
not modelled
66.0
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical cytosolic protein;PDBTitle: crystal structure of an uncharacterized protein from coxiella burnetii
34 c3hi0B_
not modelled
61.4
10
PDB header: hydrolaseChain: B: PDB Molecule: putative exopolyphosphatase;PDBTitle: crystal structure of putative exopolyphosphatase (17739545) from2 agrobacterium tumefaciens str. c58 (dupont) at 2.30 a resolution
35 c2dqbB_
not modelled
61.3
22
PDB header: hydrolase, dna binding proteinChain: B: PDB Molecule: deoxyguanosinetriphosphate triphosphohydrolase, putative;PDBTitle: crystal structure of dntp triphosphohydrolase from thermus2 thermophilus hb8, which is homologous to dgtp triphosphohydrolase
36 c2pgsA_
not modelled
54.9
29
PDB header: hydrolaseChain: A: PDB Molecule: putative deoxyguanosinetriphosphate triphosphohydrolase;PDBTitle: crystal structure of a putative deoxyguanosinetriphosphate2 triphosphohydrolase from pseudomonas syringae pv. phaseolicola 1448a
37 c3lmaC_
not modelled
44.5
19
PDB header: membrane proteinChain: C: PDB Molecule: stage v sporulation protein ad (spovad);PDBTitle: crystal structure of the stage v sporulation protein ad2 (spovad) from bacillus licheniformis. northeast structural3 genomics consortium target bir6.
38 c3hc1A_
not modelled
43.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized hdod domain protein;PDBTitle: crystal structure of hdod domain protein with unknown function2 (np_953345.1) from geobacter sulfurreducens at 1.90 a resolution
39 c3m1tA_
not modelled
41.1
14
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphohydrolase;PDBTitle: crystal structure of putative phosphohydrolase (yp_929327.1) from2 shewanella amazonensis sb2b at 1.62 a resolution
40 d1vqra_
not modelled
40.9
12
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: modified HD domain41 d1ylqa1
not modelled
38.8
30
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase42 c2w9mB_
not modelled
34.1
21
PDB header: dna replicationChain: B: PDB Molecule: polymerase x;PDBTitle: structure of family x dna polymerase from deinococcus2 radiodurans
43 d2ibna1
not modelled
31.2
16
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: MioX-like44 d2heka1
not modelled
30.7
20
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain45 c2huoA_
not modelled
26.1
26
PDB header: oxidoreductaseChain: A: PDB Molecule: inositol oxygenase;PDBTitle: crystal structure of mouse myo-inositol oxygenase in complex with2 substrate
46 d3bxda1
not modelled
26.1
26
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: MioX-like47 d1jmsa4
not modelled
21.1
17
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like48 d1wota_
not modelled
20.1
18
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase49 c3bg2A_
not modelled
19.6
23
PDB header: hydrolaseChain: A: PDB Molecule: dgtp triphosphohydrolase;PDBTitle: crystal structure of deoxyguanosinetriphosphate triphosphohydrolase2 from flavobacterium sp. med217
50 c3k1rB_
not modelled
19.4
24
PDB header: structural proteinChain: B: PDB Molecule: usher syndrome type-1g protein;PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
51 d1r89a2
not modelled
18.6
13
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Archaeal tRNA CCA-adding enzyme catalytic domain52 d2bcqa3
not modelled
17.2
10
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like53 d2b7oa1
not modelled
17.0
38
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class-II DAHP synthetase54 d1no5a_
not modelled
15.8
18
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase55 d2dula1
not modelled
15.1
23
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TRM1-like56 d1bgfa_
not modelled
12.8
22
Fold: Transcription factor STAT-4 N-domainSuperfamily: Transcription factor STAT-4 N-domainFamily: Transcription factor STAT-4 N-domain57 c3riuC_
not modelled
12.7
22
PDB header: hydrolaseChain: C: PDB Molecule: translin associated factor x, isoform b;PDBTitle: crystal structure of drosophila hexameric c3po formed by truncated2 translin and trax
58 c2bcuA_
not modelled
12.2
14
PDB header: transferase, lyase/dnaChain: A: PDB Molecule: dna polymerase lambda;PDBTitle: dna polymerase lambda in complex with a dna duplex2 containing an unpaired damp and a t:t mismatch
59 c2ihmA_
not modelled
11.8
18
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase mu;PDBTitle: polymerase mu in ternary complex with gapped 11mer dna2 duplex and bound incoming nucleotide
60 d1vola2
not modelled
11.4
17
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain61 d1es6a1
not modelled
11.3
45
Fold: EV matrix proteinSuperfamily: EV matrix proteinFamily: EV matrix protein62 d2gmwa1
not modelled
11.0
22
Fold: HAD-likeSuperfamily: HAD-likeFamily: Histidinol phosphatase-like63 c3esqA_
not modelled
11.0
22
PDB header: hydrolaseChain: A: PDB Molecule: d,d-heptose 1,7-bisphosphate phosphatase;PDBTitle: crystal structure of calcium-bound d,d-heptose 1.7-2 bisphosphate phosphatase from e. coli
64 c2r8qA_
not modelled
10.0
21
PDB header: hydrolaseChain: A: PDB Molecule: class i phosphodiesterase pdeb1;PDBTitle: structure of lmjpdeb1 in complex with ibmx
65 d1t3ta4
not modelled
9.3
44
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like66 d2fmpa3
not modelled
9.2
14
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like67 c3ndcB_
not modelled
9.2
29
PDB header: transferaseChain: B: PDB Molecule: precorrin-4 c(11)-methyltransferase;PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
68 d1vk3a1
not modelled
9.1
67
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like69 c1h2dA_
not modelled
9.1
45
PDB header: virus/viral proteinChain: A: PDB Molecule: matrix protein vp40;PDBTitle: ebola virus matrix protein vp40 n-terminal domain in2 complex with rna (low-resolution vp40[31-212] variant).
70 c2cqzA_
not modelled
9.1
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 177aa long hypothetical protein;PDBTitle: crystal structure of ph0347 protein from pyrococcus horikoshii ot3
71 d1h2ca_
not modelled
9.1
45
Fold: EV matrix proteinSuperfamily: EV matrix proteinFamily: EV matrix protein72 d1jqra_
not modelled
8.8
18
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like73 c2rffA_
not modelled
8.6
26
PDB header: transferaseChain: A: PDB Molecule: putative nucleotidyltransferase;PDBTitle: crystal structure of a putative nucleotidyltransferase2 (np_343093.1) from sulfolobus solfataricus at 1.40 a3 resolution
74 d1uqva_
not modelled
8.6
4
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain75 c1b0nB_
not modelled
7.8
32
PDB header: transcription regulatorChain: B: PDB Molecule: protein (sini protein);PDBTitle: sinr protein/sini protein complex
76 d1b0nb_
not modelled
7.8
32
Fold: Dimerisation interlockSuperfamily: SinR repressor dimerisation domain-likeFamily: SinR repressor dimerisation domain-like77 d1g60a_
not modelled
7.6
25
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Type II DNA methylase78 d1eg2a_
not modelled
7.4
17
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Type II DNA methylase79 d1vola1
not modelled
7.4
12
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain80 c3ljvA_
not modelled
7.4
9
PDB header: transcriptionChain: A: PDB Molecule: mmoq response regulator;PDBTitle: crystal structure of mmoq response regulator (fragment 29-302) from2 methylococcus capsulatus str. bath, northeast structural genomics3 consortium target mcr175m
81 c2zifB_
not modelled
7.3
25
PDB header: transferaseChain: B: PDB Molecule: putative modification methylase;PDBTitle: crystal structure of ttha0409, putative dna modification2 methylase from thermus thermophilus hb8- complexed with s-3 adenosyl-l-methionine
82 d2vana2
not modelled
7.1
21
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like83 c1xotB_
not modelled
6.3
14
PDB header: hydrolaseChain: B: PDB Molecule: camp-specific 3',5'-cyclic phosphodiesterase 4b;PDBTitle: catalytic domain of human phosphodiesterase 4b in complex with2 vardenafil
84 c2w11B_
not modelled
5.9
25
PDB header: hydrolaseChain: B: PDB Molecule: 2-haloalkanoic acid dehalogenase;PDBTitle: structure of the l-2-haloacid dehalogenase from sulfolobus2 tokodaii
85 c2ounA_
not modelled
5.9
16
PDB header: hydrolaseChain: A: PDB Molecule: camp and camp-inhibited cgmp 3',5'-cyclicPDBTitle: crystal structure of pde10a2 in complex with amp
86 c3i7aA_
not modelled
5.8
9
PDB header: hydrolaseChain: A: PDB Molecule: putative metal-dependent phosphohydrolase;PDBTitle: crystal structure of putative metal-dependent phosphohydrolase2 (yp_926882.1) from shewanella amazonensis sb2b at 2.06 a resolution
87 c1nw6A_
not modelled
5.7
17
PDB header: transferaseChain: A: PDB Molecule: modification methylase rsri;PDBTitle: structure of the beta class n6-adenine dna methyltransferase rsri2 bound to sinefungin
88 d1tbfa_
not modelled
5.7
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase89 c1xozA_
not modelled
5.7
13
PDB header: hydrolaseChain: A: PDB Molecule: cgmp-specific 3',5'-cyclic phosphodiesterase;PDBTitle: catalytic domain of human phosphodiesterase 5a in complex2 with tadalafil
90 c1es6A_
not modelled
5.6
45
PDB header: viral proteinChain: A: PDB Molecule: matrix protein vp40;PDBTitle: crystal structure of the matrix protein of ebola virus
91 d1oxja1
not modelled
5.6
12
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain92 d1sv0c_
not modelled
5.5
16
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain93 d1taza_
not modelled
5.5
31
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase94 d1pv5a_
not modelled
5.4
30
Fold: Hypothetical protein YwqGSuperfamily: Hypothetical protein YwqGFamily: Hypothetical protein YwqG95 c2zq5A_
not modelled
5.1
22
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of sulfotransferase stf1 from2 mycobacterium tuberculosis h37rv (type1 form)