| 1 | c1efpC_ |
|
 |
100.0 |
31 |
PDB header:electron transport
Chain: C: PDB Molecule:protein (electron transfer flavoprotein);
PDBTitle: electron transfer flavoprotein (etf) from paracoccus2 denitrificans
|
| 2 | d1efva2 |
|
 |
100.0 |
45 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:C-terminal domain of the electron transfer flavoprotein alpha subunit |
| 3 | d1efpa2 |
|
 |
100.0 |
48 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:C-terminal domain of the electron transfer flavoprotein alpha subunit |
| 4 | d3clsd2 |
|
 |
100.0 |
42 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:C-terminal domain of the electron transfer flavoprotein alpha subunit |
| 5 | c3ih5A_ |
|
 |
100.0 |
16 |
PDB header:electron transport
Chain: A: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: crystal structure of electron transfer flavoprotein alpha-2 subunit from bacteroides thetaiotaomicron
|
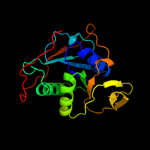
| 6 | d1efva1 |
|
 |
100.0 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
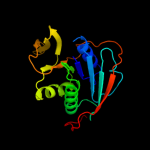
| 7 | d3clsd1 |
|
 |
100.0 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
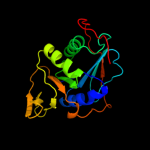
| 8 | c1t9gR_ |
|
 |
100.0 |
14 |
PDB header:oxidoreductase, electron transport
Chain: R: PDB Molecule:electron transfer flavoprotein alpha-subunit,
PDBTitle: structure of the human mcad:etf complex
|
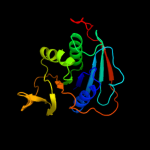
| 9 | c1o94D_ |
|
 |
100.0 |
17 |
PDB header:electron transport
Chain: D: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: ternary complex between trimethylamine dehydrogenase and2 electron transferring flavoprotein
|
| 10 | d1efpa1 |
|
 |
100.0 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 11 | c3fetA_ |
|
 |
100.0 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:electron transfer flavoprotein subunit alpha related
PDBTitle: crystal structure of the electron transfer flavoprotein subunit alpha2 related protein ta0212 from thermoplasma acidophilum
|
| 12 | d1efvb_ |
|
 |
99.8 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 13 | d3clsc1 |
|
 |
99.8 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 14 | d1efpb_ |
|
 |
99.8 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 15 | d1o94c_ |
|
 |
99.7 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 16 | d1t9ba1 |
|
 |
98.9 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 17 | d1ozha1 |
|
 |
98.8 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 18 | d2ez9a1 |
|
 |
98.5 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 19 | d2ji7a1 |
|
 |
98.4 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 20 | d2ihta1 |
|
 |
98.3 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 21 | d1q6za1 |
|
not modelled |
98.0 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 22 | c1ozhD_ |
|
not modelled |
98.0 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:acetolactate synthase, catabolic;
PDBTitle: the crystal structure of klebsiella pneumoniae acetolactate2 synthase with enzyme-bound cofactor and with an unusual3 intermediate.
|
| 23 | d1ybha1 |
|
not modelled |
97.8 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 24 | c2q27B_ |
|
not modelled |
97.7 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:oxalyl-coa decarboxylase;
PDBTitle: crystal structure of oxalyl-coa decarboxylase from escherichia coli
|
| 25 | d2djia1 |
|
not modelled |
97.6 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 26 | c1powA_ |
|
not modelled |
97.6 |
22 |
PDB header:oxidoreductase(oxygen as acceptor)
Chain: A: PDB Molecule:pyruvate oxidase;
PDBTitle: the refined structures of a stabilized mutant and of wild-type2 pyruvate oxidase from lactobacillus plantarum
|
| 27 | c2pgnA_ |
|
not modelled |
97.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:cyclohexane-1,2-dione hydrolase (cdh);
PDBTitle: the crystal structure of fad and thdp-dependent cyclohexane-1,2-dione2 hydrolase in complex with cyclohexane-1,2-dione
|
| 28 | c2djiA_ |
|
not modelled |
97.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate oxidase;
PDBTitle: crystal structure of pyruvate oxidase from aerococcus2 viridans containing fad
|
| 29 | c3lq1A_ |
|
not modelled |
97.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-
PDBTitle: crystal structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene2 1-carboxylic acid synthase/2-oxoglutarate decarboxylase3 from listeria monocytogenes str. 4b f2365
|
| 30 | c2ji6B_ |
|
not modelled |
97.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:oxalyl-coa decarboxylase;
PDBTitle: x-ray structure of oxalyl-coa decarboxylase in complex with2 3-deaza-thdp and oxalyl-coa
|
| 31 | c1t9dB_ |
|
not modelled |
97.3 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:acetolactate synthase, mitochondrial;
PDBTitle: crystal structure of yeast acetohydroxyacid synthase in2 complex with a sulfonylurea herbicide, metsulfuron methyl
|
| 32 | c1upaC_ |
|
not modelled |
97.1 |
14 |
PDB header:synthase
Chain: C: PDB Molecule:carboxyethylarginine synthase;
PDBTitle: carboxyethylarginine synthase from streptomyces2 clavuligerus (semet structure)
|
| 33 | c1yi1A_ |
|
not modelled |
97.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase;
PDBTitle: crystal structure of arabidopsis thaliana acetohydroxyacid synthase in2 complex with a sulfonylurea herbicide, tribenuron methyl
|
| 34 | c2ag1A_ |
|
not modelled |
97.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:benzaldehyde lyase;
PDBTitle: crystal structure of benzaldehyde lyase (bal)- semet
|
| 35 | c3eyaE_ |
|
not modelled |
96.9 |
16 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:pyruvate dehydrogenase [cytochrome];
PDBTitle: structural basis for membrane binding and catalytic2 activation of the peripheral membrane enzyme pyruvate3 oxidase from escherichia coli
|
| 36 | c2panF_ |
|
not modelled |
96.7 |
20 |
PDB header:lyase
Chain: F: PDB Molecule:glyoxylate carboligase;
PDBTitle: crystal structure of e. coli glyoxylate carboligase
|
| 37 | c2v3wC_ |
|
not modelled |
96.6 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:benzoylformate decarboxylase;
PDBTitle: crystal structure of the benzoylformate decarboxylase2 variant l461a from pseudomonas putida
|
| 38 | c2x7jA_ |
|
not modelled |
96.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: structure of the menaquinone biosynthesis protein mend from2 bacillus subtilis
|
| 39 | d1zpda1 |
|
not modelled |
96.5 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 40 | d1pvda1 |
|
not modelled |
96.2 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 41 | d1m2ka_ |
|
not modelled |
96.1 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 42 | d1yc5a1 |
|
not modelled |
96.0 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 43 | c1jscA_ |
|
not modelled |
95.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:acetohydroxy-acid synthase;
PDBTitle: crystal structure of the catalytic subunit of yeast2 acetohydroxyacid synthase: a target for herbicidal3 inhibitors
|
| 44 | d2b4ya1 |
|
not modelled |
95.4 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 45 | c3glsC_ |
|
not modelled |
95.2 |
27 |
PDB header:hydrolase
Chain: C: PDB Molecule:nad-dependent deacetylase sirtuin-3,
PDBTitle: crystal structure of human sirt3
|
| 46 | c3k35D_ |
|
not modelled |
95.1 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: crystal structure of human sirt6
|
| 47 | c1zpdA_ |
|
not modelled |
94.9 |
14 |
PDB header:alcohol fermentation
Chain: A: PDB Molecule:pyruvate decarboxylase;
PDBTitle: pyruvate decarboxylase from zymomonas mobilis
|
| 48 | d1ma3a_ |
|
not modelled |
94.9 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 49 | c3pkiF_ |
|
not modelled |
94.5 |
20 |
PDB header:hydrolase
Chain: F: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: human sirt6 crystal structure in complex with adp ribose
|
| 50 | c2vbiF_ |
|
not modelled |
94.4 |
11 |
PDB header:lyase
Chain: F: PDB Molecule:pyruvate decarboxylase;
PDBTitle: holostructure of pyruvate decarboxylase from acetobacter2 pasteurianus
|
| 51 | c2jlaD_ |
|
not modelled |
94.3 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: crystal structure of e.coli mend, 2-succinyl-5-enolpyruvyl-2 6-hydroxy-3-cyclohexadiene-1-carboxylate synthase - semet3 protein
|
| 52 | d1q1aa_ |
|
not modelled |
94.3 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 53 | c1q14A_ |
|
not modelled |
93.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:hst2 protein;
PDBTitle: structure and autoregulation of the yeast hst2 homolog of sir2
|
| 54 | d1ovma1 |
|
not modelled |
93.2 |
12 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 55 | c2w93A_ |
|
not modelled |
92.7 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:pyruvate decarboxylase isozyme 1;
PDBTitle: crystal structure of the saccharomyces cerevisiae pyruvate2 decarboxylase variant e477q in complex with the surrogate3 pyruvamide
|
| 56 | d1s5pa_ |
|
not modelled |
92.5 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 57 | d1j8fa_ |
|
not modelled |
91.7 |
32 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 58 | c3jwpA_ |
|
not modelled |
90.7 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein sir2 homologue;
PDBTitle: crystal structure of plasmodium falciparum sir2a (pf13_0152) in2 complex with amp
|
| 59 | c2hjhB_ |
|
not modelled |
90.0 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:nad-dependent histone deacetylase sir2;
PDBTitle: crystal structure of the sir2 deacetylase
|
| 60 | c1ovmC_ |
|
not modelled |
89.6 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:indole-3-pyruvate decarboxylase;
PDBTitle: crystal structure of indolepyruvate decarboxylase from2 enterobacter cloacae
|
| 61 | c2vbgB_ |
|
not modelled |
88.9 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:branched-chain alpha-ketoacid decarboxylase;
PDBTitle: the complex structure of the branched-chain keto acid2 decarboxylase (kdca) from lactococcus lactis with 2r-1-3 hydroxyethyl-deazathdp
|
| 62 | c2nxwB_ |
|
not modelled |
87.5 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:phenyl-3-pyruvate decarboxylase;
PDBTitle: crystal structure of phenylpyruvate decarboxylase of azospirillum2 brasilense
|
| 63 | c3cf4G_ |
|
not modelled |
84.7 |
21 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:acetyl-coa decarboxylase/synthase epsilon subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
|
| 64 | d1uana_ |
|
not modelled |
83.4 |
13 |
Fold:LmbE-like
Superfamily:LmbE-like
Family:LmbE-like |
| 65 | d1tq8a_ |
|
not modelled |
82.7 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 66 | d1x94a_ |
|
not modelled |
78.7 |
16 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 67 | c2x3yA_ |
|
not modelled |
77.3 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoheptose isomerase;
PDBTitle: crystal structure of gmha from burkholderia pseudomallei
|
| 68 | c2ixdB_ |
|
not modelled |
70.3 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmbe-related protein;
PDBTitle: crystal structure of the putative deacetylase bc1534 from2 bacilus cereus
|
| 69 | c2pn1A_ |
|
not modelled |
67.2 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoylphosphate synthase large subunit;
PDBTitle: crystal structure of carbamoylphosphate synthase large subunit (split2 gene in mj) (zp_00538348.1) from exiguobacterium sp. 255-15 at 2.00 a3 resolution
|
| 70 | c3fxaA_ |
|
not modelled |
59.9 |
14 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sis domain protein;
PDBTitle: crystal structure of a putative sugar-phosphate isomerase2 (lmof2365_0531) from listeria monocytogenes str. 4b f2365 at 1.60 a3 resolution
|
| 71 | c2ze5A_ |
|
not modelled |
53.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:isopentenyl transferase;
PDBTitle: crystal structure of adenosine phosphate-isopentenyltransferase
|
| 72 | c1nvmG_ |
|
not modelled |
52.7 |
12 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 73 | c3shoA_ |
|
not modelled |
52.0 |
21 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, rpir family;
PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
|
| 74 | d1tk9a_ |
|
not modelled |
51.7 |
14 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 75 | c2xhzC_ |
|
not modelled |
49.5 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:arabinose 5-phosphate isomerase;
PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
|
| 76 | c1nriA_ |
|
not modelled |
46.1 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0754;
PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
|
| 77 | d1nria_ |
|
not modelled |
46.1 |
13 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 78 | c3d3qB_ |
|
not modelled |
45.1 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: crystal structure of trna delta(2)-isopentenylpyrophosphate2 transferase (se0981) from staphylococcus epidermidis.3 northeast structural genomics consortium target ser100
|
| 79 | c3dahB_ |
|
not modelled |
44.4 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
|
| 80 | d1moqa_ |
|
not modelled |
44.1 |
13 |
Fold:SIS domain
Superfamily:SIS domain
Family:double-SIS domain |
| 81 | c2dwcB_ |
|
not modelled |
44.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
| 82 | c3uvzB_ |
|
not modelled |
42.5 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase, atpase subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
|
| 83 | d2iyva1 |
|
not modelled |
42.0 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 84 | c2amlB_ |
|
not modelled |
41.3 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:sis domain protein;
PDBTitle: crystal structure of lmo0035 protein (46906266) from listeria2 monocytogenes 4b f2365 at 1.50 a resolution
|
| 85 | d1kjqa2 |
|
not modelled |
40.9 |
17 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 86 | c3crqA_ |
|
not modelled |
40.1 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: structure of trna dimethylallyltransferase: rna2 modification through a channel
|
| 87 | c3exaD_ |
|
not modelled |
40.0 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: crystal structure of the full-length trna2 isopentenylpyrophosphate transferase (bh2366) from3 bacillus halodurans, northeast structural genomics4 consortium target bhr41.
|
| 88 | c1u9yD_ |
|
not modelled |
38.1 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
|
| 89 | c3ga2A_ |
|
not modelled |
36.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of the endonuclease_v (bsu36170) from2 bacillus subtilis, northeast structural genomics3 consortium target sr624
|
| 90 | c3a8tA_ |
|
not modelled |
35.1 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate isopentenyltransferase;
PDBTitle: plant adenylate isopentenyltransferase in complex with atp
|
| 91 | c1dkrB_ |
|
not modelled |
33.6 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
| 92 | c3euaD_ |
|
not modelled |
32.9 |
29 |
PDB header:isomerase
Chain: D: PDB Molecule:putative fructose-aminoacid-6-phosphate deglycase;
PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
|
| 93 | d1jmva_ |
|
not modelled |
32.8 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 94 | c3bo9B_ |
|
not modelled |
30.8 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nitroalkan dioxygenase;
PDBTitle: crystal structure of putative nitroalkan dioxygenase (tm0800) from2 thermotoga maritima at 2.71 a resolution
|
| 95 | c3t61A_ |
|
not modelled |
30.8 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:gluconokinase;
PDBTitle: crystal structure of a gluconokinase from sinorhizobium meliloti 1021
|
| 96 | d1q77a_ |
|
not modelled |
30.5 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 97 | d1kaga_ |
|
not modelled |
30.4 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 98 | c3trjC_ |
|
not modelled |
29.7 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
| 99 | c3fozB_ |
|
not modelled |
28.9 |
27 |
PDB header:transferase/rna
Chain: B: PDB Molecule:trna delta(2)-isopentenylpyrophosphate transferase;
PDBTitle: structure of e. coli isopentenyl-trna transferase in complex with e.2 coli trna(phe)
|
| 100 | c3m0zD_ |
|
not modelled |
28.7 |
22 |
PDB header:lyase
Chain: D: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of putative aldolase from klebsiella2 pneumoniae.
|
| 101 | d1a9xa4 |
|
not modelled |
28.5 |
9 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 102 | d1nvma2 |
|
not modelled |
27.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 103 | c3tbfA_ |
|
not modelled |
27.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:glucosamine--fructose-6-phosphate aminotransferase
PDBTitle: c-terminal domain of glucosamine-fructose-6-phosphate aminotransferase2 from francisella tularensis.
|
| 104 | c2p3wB_ |
|
not modelled |
25.8 |
21 |
PDB header:protein binding
Chain: B: PDB Molecule:probable serine protease htra3;
PDBTitle: crystal structure of the htra3 pdz domain bound to a phage-derived2 ligand (fgrwv)
|
| 105 | d1pama3 |
|
not modelled |
25.5 |
31 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 106 | d1nt2a_ |
|
not modelled |
25.4 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Fibrillarin homologue |
| 107 | c3eplA_ |
|
not modelled |
24.8 |
26 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna isopentenyltransferase;
PDBTitle: crystallographic snapshots of eukaryotic2 dimethylallyltransferase acting on trna: insight into trna3 recognition and reaction mechanism
|
| 108 | c1rr2A_ |
|
not modelled |
24.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 109 | d1rkba_ |
|
not modelled |
24.1 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 110 | c3be4A_ |
|
not modelled |
23.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of cryptosporidium parvum adenylate kinase cgd5_3360
|
| 111 | c3lpnB_ |
|
not modelled |
23.5 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
|
| 112 | d1cxla3 |
|
not modelled |
22.1 |
38 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 113 | c3muxB_ |
|
not modelled |
21.8 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:putative 4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
|
| 114 | c3m6yA_ |
|
not modelled |
21.4 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
|
| 115 | c2k1hA_ |
|
not modelled |
21.4 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ser13;
PDBTitle: solution nmr structure of ser13 from staphylococcus epidermidis.2 northeast structural genomics consortium target ser13
|
| 116 | d1g8sa_ |
|
not modelled |
20.7 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Fibrillarin homologue |

