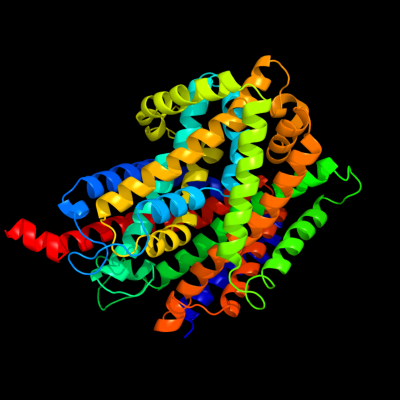
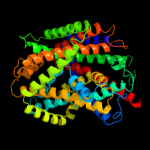
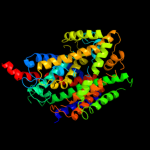
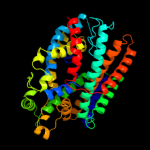
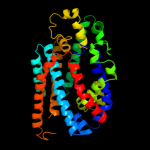
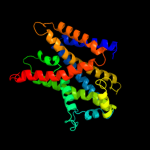
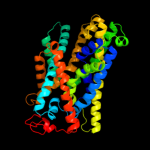
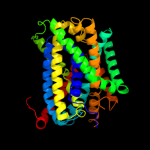
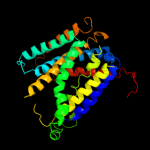
1 c3hfxA_
100.0
24
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
2 c2w8aC_
100.0
34
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
3 c2xq2A_
98.5
15
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
4 c3dh4A_
97.9
13
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
5 d2a65a1
97.7
12
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like6 c2jlnA_
97.5
10
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
7 c3giaA_
96.9
12
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
8 c3lrcC_
94.1
14
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
9 c1u7mB_
50.3
25
PDB header: de novo proteinChain: B: PDB Molecule: four-helix bundle model;PDBTitle: solution structure of a diiron protein model: due ferri(ii)2 turn mutant
10 d1ylxa1
36.8
22
Fold: N domain of copper amine oxidase-likeSuperfamily: GK1464-likeFamily: GK1464-like11 c2hz8A_
36.3
27
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed diiron protein;PDBTitle: qm/mm structure refined from nmr-structure of a single2 chain diiron protein
12 c1lt1G_
32.4
33
PDB header: de novo proteinChain: G: PDB Molecule: l13g-df1;PDBTitle: sliding helix induced change of coordination geometry in a2 model di-mn(ii) protein
13 d1z2la1
25.1
9
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases14 d2cv4a1
25.0
10
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like15 c3gztF_
22.4
30
PDB header: virusChain: F: PDB Molecule: outer capsid glycoprotein vp7;PDBTitle: vp7 recoated rotavirus dlp
16 c3s0xB_
22.4
12
PDB header: hydrolaseChain: B: PDB Molecule: peptidase a24b, flak domain protein;PDBTitle: the crystal structure of gxgd membrane protease flak
17 c2kvlA_
20.6
33
PDB header: viral proteinChain: A: PDB Molecule: major outer capsid protein vp7;PDBTitle: nmr structure of the c-terminal domain of vp7
18 d2axtm1
19.8
31
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein M, PsbMFamily: PsbM-like19 d1j5ya2
19.0
12
Fold: HPr-likeSuperfamily: Putative transcriptional regulator TM1602, C-terminal domainFamily: Putative transcriptional regulator TM1602, C-terminal domain20 d2guka1
17.6
45
Fold: PG1857-likeSuperfamily: PG1857-likeFamily: PG1857-like21 c3g40A_
not modelled
16.7
16
PDB header: transport proteinChain: A: PDB Molecule: na-k-cl cotransporter;PDBTitle: crystal structure of the cytoplasmic domain of a2 prokaryotic cation chloride cotransporter
22 d1kf6d_
not modelled
15.6
18
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)23 c1x59A_
not modelled
15.2
60
PDB header: protein bindingChain: A: PDB Molecule: histidyl-trna synthetase;PDBTitle: solution structures of the whep-trs domain of human2 histidyl-trna synthetase
24 c3l09B_
not modelled
14.8
19
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator2 (jann_22dec04_contig27_revised_gene3569) from jannaschia sp. ccs1 at3 2.81 a resolution
25 c1bmlD_
not modelled
14.8
16
PDB header: blood clottingChain: D: PDB Molecule: streptokinase;PDBTitle: complex of the catalytic domain of human plasmin and streptokinase
26 d1c4pc_
not modelled
13.7
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase27 c2qksA_
not modelled
13.5
16
PDB header: metal transportChain: A: PDB Molecule: kir3.1-prokaryotic kir channel chimera;PDBTitle: crystal structure of a kir3.1-prokaryotic kir channel chimera
28 c3g36D_
not modelled
13.4
13
PDB header: nuclear proteinChain: D: PDB Molecule: protein dpy-30 homolog;PDBTitle: crystal structure of the human dpy-30-like c-terminal domain
29 c3u4gA_
not modelled
13.3
37
PDB header: transferaseChain: A: PDB Molecule: namn:dmb phosphoribosyltransferase;PDBTitle: the structure of cobt from pyrococcus horikoshii
30 d1a7ja_
not modelled
13.2
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase31 d2f2ha1
not modelled
12.8
25
Fold: Putative glucosidase YicI, C-terminal domainSuperfamily: Putative glucosidase YicI, C-terminal domainFamily: Putative glucosidase YicI, C-terminal domain32 d2p6va1
not modelled
12.7
24
Fold: TAFH domain-likeSuperfamily: TAFH domain-likeFamily: TAFH domain-like33 d2vo9a1
not modelled
11.7
12
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanY-like34 c2z9oB_
not modelled
10.6
6
PDB header: replication/dnaChain: B: PDB Molecule: replication initiation protein;PDBTitle: crystal structure of the dimeric form of repe in complex with the repe2 operator dna
35 c1z0zC_
not modelled
10.0
17
PDB header: transferaseChain: C: PDB Molecule: probable inorganic polyphosphate/atp-nad kinase;PDBTitle: crystal structure of a nad kinase from archaeoglobus2 fulgidus in complex with nad
36 d1s0yb_
not modelled
10.0
36
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like37 d2g3qa1
not modelled
9.7
20
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain38 c3ej9D_
not modelled
9.7
36
PDB header: hydrolaseChain: D: PDB Molecule: beta-subunit of trans-3-chloroacrylic acid dehalogenase;PDBTitle: structural and mechanistic analysis of trans-3-chloroacrylic acid2 dehalogenase activity
39 c2ksnA_
not modelled
9.5
15
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin domain-containing protein 2;PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
40 c3eofB_
not modelled
9.3
21
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of putative oxidoreductase (yp_213212.1) from2 bacteroides fragilis nctc 9343 at 1.99 a resolution
41 d1z0sa1
not modelled
9.0
17
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: NAD kinase-like42 c3kl9F_
not modelled
8.9
12
PDB header: hydrolaseChain: F: PDB Molecule: glutamyl aminopeptidase;PDBTitle: crystal structure of pepa from streptococcus pneumoniae
43 c2dn9A_
not modelled
8.9
9
PDB header: apoptosis, chaperoneChain: A: PDB Molecule: dnaj homolog subfamily a member 3;PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
44 c3zqsB_
not modelled
8.9
8
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase fancl;PDBTitle: human fancl central domain
45 c3by7E_
not modelled
8.8
50
PDB header: unknown functionChain: E: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein structurally similar to sm/lsm-like2 rna-binding proteins (jcvi_pep_1096686650277) from uncultured marine3 organism at 2.60 a resolution
46 d1gxma_
not modelled
8.7
28
Fold: alpha/alpha toroidSuperfamily: Family 10 polysaccharide lyaseFamily: Family 10 polysaccharide lyase47 d1f9fa_
not modelled
8.6
16
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain48 c2la3A_
not modelled
8.3
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr structure of the protein np_344798.1 reveals a cca-adding2 enzyme head domain
49 d1repc2
not modelled
8.3
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Replication initiation protein50 c3lklB_
not modelled
8.2
19
PDB header: transport proteinChain: B: PDB Molecule: antisigma-factor antagonist stas;PDBTitle: crystal structure of the c-terminal domain of anti-sigma factor2 antagonist stas from rhodobacter sphaeroides
51 c2vo9C_
not modelled
8.1
12
PDB header: hydrolaseChain: C: PDB Molecule: l-alanyl-d-glutamate peptidase;PDBTitle: crystal structure of the enzymatically active domain of the2 listeria monocytogenes bacteriophage 500 endolysin ply500
52 c1pk1B_
not modelled
8.1
10
PDB header: transcription repressionChain: B: PDB Molecule: sex comb on midleg cg9495-pa;PDBTitle: hetero sam domain structure of ph and scm.
53 d2a7sa1
not modelled
8.1
19
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain54 d3b45a1
not modelled
8.0
20
Fold: Rhomboid-likeSuperfamily: Rhomboid-likeFamily: Rhomboid-like55 c2jnvA_
not modelled
7.9
6
PDB header: metal transportChain: A: PDB Molecule: nifu-like protein 1, chloroplast;PDBTitle: solution structure of c-terminal domain of nifu-like2 protein from oryza sativa
56 c3rzaA_
not modelled
7.9
8
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidase;PDBTitle: crystal structure of a tripeptidase (sav1512) from staphylococcus2 aureus subsp. aureus mu50 at 2.10 a resolution
57 d1y0ua_
not modelled
7.9
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators58 d1o5wa2
not modelled
7.8
9
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase59 c2l69A_
not modelled
7.8
29
PDB header: de novo proteinChain: A: PDB Molecule: rossmann 2x3 fold protein;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
60 c3jycA_
not modelled
7.7
21
PDB header: metal transportChain: A: PDB Molecule: inward-rectifier k+ channel kir2.2;PDBTitle: crystal structure of the eukaryotic strong inward-rectifier2 k+ channel kir2.2 at 3.1 angstrom resolution
61 d1r76a_
not modelled
7.7
18
Fold: alpha/alpha toroidSuperfamily: Family 10 polysaccharide lyaseFamily: Family 10 polysaccharide lyase62 d1vrga1
not modelled
7.7
19
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain63 c3ps5A_
not modelled
7.7
10
PDB header: hydrolase, signaling proteinChain: A: PDB Molecule: tyrosine-protein phosphatase non-receptor type 6;PDBTitle: crystal structure of the full-length human protein tyrosine2 phosphatase shp-1
64 d1z3xa1
not modelled
7.6
20
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: GUN4-associated domain65 c3gycB_
not modelled
7.4
16
PDB header: hydrolaseChain: B: PDB Molecule: putative glycoside hydrolase;PDBTitle: crystal structure of putative glycoside hydrolase (yp_001304622.1)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
66 c3c4rC_
not modelled
7.4
11
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
67 d1qqra_
not modelled
7.4
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase68 d2j8cl1
not modelled
7.3
14
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits69 c2k10A_
not modelled
7.3
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: ranatuerin-2csa;PDBTitle: confirmational analysis of the broad-spectrum antibacterial2 peptide, rantuerin-2csa: identification of a full length3 helix-turn-helix motif
70 c3kv1A_
not modelled
7.2
9
PDB header: transcriptionChain: A: PDB Molecule: transcriptional repressor;PDBTitle: crystal structure of putative sugar-binding domain of transcriptional2 repressor from vibrio fischeri
71 d1pixa2
not modelled
7.2
11
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain72 d1egwa_
not modelled
7.1
10
Fold: SRF-likeSuperfamily: SRF-likeFamily: SRF-like73 c2bj3D_
not modelled
7.1
20
PDB header: transcriptionChain: D: PDB Molecule: nickel responsive regulator;PDBTitle: nikr-apo
74 c3au5B_
not modelled
7.0
18
PDB header: motor proteinChain: B: PDB Molecule: myosin-x;PDBTitle: structure of the human myosin-x myth4-ferm cassette
75 d1nj1a1
not modelled
7.0
12
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS76 d1rh5a_
not modelled
7.0
13
Fold: Preprotein translocase SecY subunitSuperfamily: Preprotein translocase SecY subunitFamily: Preprotein translocase SecY subunit77 c1v85A_
not modelled
7.0
10
PDB header: apoptosisChain: A: PDB Molecule: similar to ring finger protein 36;PDBTitle: sterile alpha motif (sam) domain of mouse bifunctional2 apoptosis regulator
78 d1pk3a1
not modelled
7.0
10
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain79 d1udxa3
not modelled
7.0
17
Fold: Obg GTP-binding protein C-terminal domainSuperfamily: Obg GTP-binding protein C-terminal domainFamily: Obg GTP-binding protein C-terminal domain80 d2hzaa1
not modelled
7.0
27
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like81 c3kpaB_
not modelled
7.0
33
PDB header: ligaseChain: B: PDB Molecule: probable ubiquitin fold modifier conjugating enzyme;PDBTitle: ubiquitin fold modifier conjugating enzyme from leishmania major2 (probable)
82 d2p41a1
not modelled
6.9
27
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: mRNA cap methylase83 c2o9xA_
not modelled
6.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: reductase, assembly protein;PDBTitle: crystal structure of a putative redox enzyme maturation protein from2 archaeoglobus fulgidus
84 c1q5vB_
not modelled
6.9
33
PDB header: transcriptionChain: B: PDB Molecule: nickel responsive regulator;PDBTitle: apo-nikr
85 c1usdA_
not modelled
6.8
35
PDB header: signaling proteinChain: A: PDB Molecule: vasodilator-stimulated phosphoprotein;PDBTitle: human vasp tetramerisation domain l352m
86 c3bm2B_
not modelled
6.8
9
PDB header: oxidoreductaseChain: B: PDB Molecule: protein ydja;PDBTitle: crystal structure of a minimal nitroreductase ydja from escherichia2 coli k12 with and without fmn cofactor
87 c3eyyA_
not modelled
6.8
16
PDB header: transportChain: A: PDB Molecule: putative iron uptake regulatory protein;PDBTitle: structural basis for the specialization of nur, a nickel-2 specific fur homologue, in metal sensing and dna3 recognition
88 c3k7cC_
not modelled
6.8
10
PDB header: protein bindingChain: C: PDB Molecule: putative ntf2-like transpeptidase;PDBTitle: crystal structure of putative ntf2-like transpeptidase (np_281412.1)2 from campylobacter jejuni at 2.00 a resolution
89 c3qyfA_
not modelled
6.8
15
PDB header: antiviral proteinChain: A: PDB Molecule: crispr-associated protein;PDBTitle: crystal structure of the crispr-associated protein sso1393 from2 sulfolobus solfataricus
90 c3komB_
not modelled
6.8
9
PDB header: transferaseChain: B: PDB Molecule: transketolase;PDBTitle: crystal structure of apo transketolase from francisella tularensis
91 d1ej5a_
not modelled
6.7
25
Fold: Wiscott-Aldrich syndrome protein, WASP, C-terminal domainSuperfamily: Wiscott-Aldrich syndrome protein, WASP, C-terminal domainFamily: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain92 c3o5cA_
not modelled
6.7
10
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c551 peroxidase;PDBTitle: cytochrome c peroxidase bccp of shewanella oneidensis
93 d1y5ic1
not modelled
6.7
9
Fold: Heme-binding four-helical bundleSuperfamily: Respiratory nitrate reductase 1 gamma chainFamily: Respiratory nitrate reductase 1 gamma chain94 d2otaa1
not modelled
6.7
36
Fold: YejL-likeSuperfamily: YejL-likeFamily: YejL-like95 c3lloA_
not modelled
6.7
22
PDB header: motor proteinChain: A: PDB Molecule: prestin;PDBTitle: crystal structure of the stas domain of motor protein prestin (anion2 transporter slc26a5)
96 c3ng0A_
not modelled
6.7
8
PDB header: ligaseChain: A: PDB Molecule: glutamine synthetase;PDBTitle: crystal structure of glutamine synthetase from synechocystis sp. pcc2 6803
97 d2hzab1
not modelled
6.6
27
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like98 d1ji5a_
not modelled
6.6
17
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin99 c2ca9B_
not modelled
6.6
20
PDB header: transcriptional regulationChain: B: PDB Molecule: putative nickel-responsive regulator;PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation