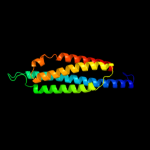
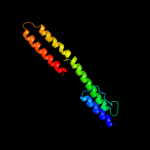
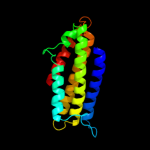
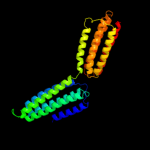
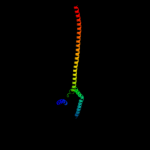
1 c1qu7A_
99.9
99
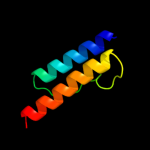
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: four helical-bundle structure of the cytoplasmic domain of a serine2 chemotaxis receptor
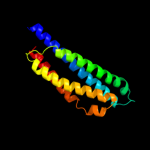
2 c2ch7A_
99.7
26
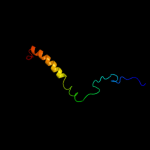
PDB header: chemotaxisChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
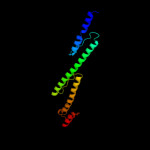
3 c3g67A_
99.4
20
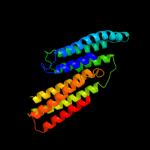
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
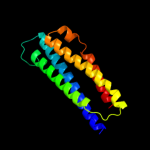
4 c2d4uA_
98.9
99
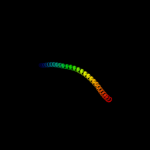
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: crystal structure of the ligand binding domain of the bacterial serine2 chemoreceptor tsr
5 d2liga_
98.2
36
Fold: Four-helical up-and-down bundleSuperfamily: Aspartate receptor, ligand-binding domainFamily: Aspartate receptor, ligand-binding domain6 d2asra_
97.8
35
Fold: Four-helical up-and-down bundleSuperfamily: Aspartate receptor, ligand-binding domainFamily: Aspartate receptor, ligand-binding domain7 c3lnrA_
97.7
13
PDB header: signaling proteinChain: A: PDB Molecule: aerotaxis transducer aer2;PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
8 d1vlta_
97.4
36
Fold: Four-helical up-and-down bundleSuperfamily: Aspartate receptor, ligand-binding domainFamily: Aspartate receptor, ligand-binding domain9 d2asxa1
96.8
23
Fold: HAMP domain-likeSuperfamily: HAMP domain-likeFamily: HAMP domain10 c2rm8A_
96.4
20
PDB header: signaling proteinChain: A: PDB Molecule: sensory rhodopsin ii transducer;PDBTitle: the solution structure of phototactic transducer protein2 htrii linker region from natronomonas pharaonis
11 c1sj8A_
94.3
11
PDB header: structural proteinChain: A: PDB Molecule: talin 1;PDBTitle: crystal structure of talin residues 482-789
12 c2wpqA_
94.2
7
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
13 c3zrwB_
93.6
18
PDB header: signaling proteinChain: B: PDB Molecule: af1503 protein, osmolarity sensor protein envz;PDBTitle: the structure of the dimeric hamp-dhp fusion a291v mutant
14 c2kbbA_
77.1
13
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: nmr structure of the talin rod domain, 1655-1822
15 c3dyjA_
75.8
8
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: crystal structure a talin rod fragment
16 c2qihA_
73.5
12
PDB header: cell adhesionChain: A: PDB Molecule: protein uspa1;PDBTitle: crystal structure of 527-665 fragment of uspa1 protein from2 moraxella catarrhalis
17 c1deqF_
68.5
8
PDB header: PDB COMPND: 18 c3ojaB_
68.5
9
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
19 c1ei3C_
63.3
6
PDB header: PDB COMPND: 20 c1deqO_
61.1
16
PDB header: PDB COMPND: 21 c3gvmA_
not modelled
56.3
14
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
22 c1ei3E_
not modelled
56.0
12
PDB header: PDB COMPND: 23 c2vs0B_
not modelled
50.2
14
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
24 c1urqA_
not modelled
43.6
19
PDB header: transport proteinChain: A: PDB Molecule: m-tomosyn isoform;PDBTitle: crystal structure of neuronal q-snares in complex with2 r-snare motif of tomosyn
25 c3ipdB_
not modelled
39.1
10
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
26 c3b5nF_
not modelled
37.4
12
PDB header: membrane proteinChain: F: PDB Molecule: protein sso1;PDBTitle: structure of the yeast plasma membrane snare complex
27 c2kseA_
not modelled
34.0
10
PDB header: transferaseChain: A: PDB Molecule: sensor protein qsec;PDBTitle: backbone structure of the membrane domain of e. coli2 histidine kinase receptor qsec, center for structures of3 membrane proteins (csmp) target 4311c
28 c2ieqC_
not modelled
32.8
14
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: core structure of s2 from the human coronavirus nl63 spike2 glycoprotein
29 c1sfcJ_
not modelled
32.0
10
PDB header: transport proteinChain: J: PDB Molecule: protein (syntaxin 1a);PDBTitle: neuronal synaptic fusion complex
30 c1n7sB_
not modelled
30.2
11
PDB header: transport proteinChain: B: PDB Molecule: syntaxin 1a;PDBTitle: high resolution structure of a truncated neuronal snare2 complex
31 c3ok8A_
not modelled
29.3
8
PDB header: protein bindingChain: A: PDB Molecule: brain-specific angiogenesis inhibitor 1-associated proteinPDBTitle: i-bar of pinkbar
32 c1i49A_
not modelled
25.3
16
PDB header: signaling proteinChain: A: PDB Molecule: arfaptin 2;PDBTitle: crystal structure analysis of arfaptin
33 c3hd7A_
not modelled
24.9
22
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
34 c2npsB_
not modelled
23.5
14
PDB header: transport proteinChain: B: PDB Molecule: syntaxin 13;PDBTitle: crystal structure of the early endosomal snare complex
35 c1sfcI_
not modelled
21.6
21
PDB header: transport proteinChain: I: PDB Molecule: protein (synaptobrevin 2);PDBTitle: neuronal synaptic fusion complex
36 c1nafA_
not modelled
21.3
15
PDB header: signaling protein, membrane proteinChain: A: PDB Molecule: adp-ribosylation factor binding protein gga1;PDBTitle: crystal structure of the human gga1 gat domain
37 d1eq1a_
not modelled
20.5
11
Fold: Apolipophorin-IIISuperfamily: Apolipophorin-IIIFamily: Apolipophorin-III38 c3b5nE_
not modelled
19.3
15
PDB header: membrane proteinChain: E: PDB Molecule: synaptobrevin homolog 1;PDBTitle: structure of the yeast plasma membrane snare complex
39 c2d4yA_
not modelled
19.2
10
PDB header: structural proteinChain: A: PDB Molecule: flagellar hook-associated protein 1;PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
40 c2bezC_
not modelled
19.2
14
PDB header: viral proteinChain: C: PDB Molecule: e2 glycoprotein;PDBTitle: structure of a proteolitically resistant core from the2 severe acute respiratory syndrome coronavirus s2 fusion3 protein
41 c1gl2A_
not modelled
18.9
13
PDB header: membrane proteinChain: A: PDB Molecule: endobrevin;PDBTitle: crystal structure of an endosomal snare core complex
42 c2npsA_
not modelled
18.0
26
PDB header: transport proteinChain: A: PDB Molecule: vesicle-associated membrane protein 4;PDBTitle: crystal structure of the early endosomal snare complex
43 c1n7sA_
not modelled
15.9
22
PDB header: transport proteinChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: high resolution structure of a truncated neuronal snare2 complex
44 c3ghgK_
not modelled
15.9
9
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
45 c1zvaA_
not modelled
14.5
14
PDB header: viral proteinChain: A: PDB Molecule: e2 glycoprotein;PDBTitle: a structure-based mechanism of sars virus membrane fusion
46 d1wa8a1
not modelled
13.6
11
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like47 c1bf5A_
not modelled
13.4
7
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
48 d1oxza_
not modelled
13.0
14
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain49 c1oxzA_
not modelled
13.0
14
PDB header: membrane proteinChain: A: PDB Molecule: adp-ribosylation factor binding protein gga1;PDBTitle: crystal structure of the human gga1 gat domain
50 d1wr6a1
not modelled
12.6
8
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain51 c1kmiZ_
not modelled
12.5
10
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
52 d1ez3a_
not modelled
12.3
10
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins53 c2d3eD_
not modelled
12.2
10
PDB header: contractile proteinChain: D: PDB Molecule: general control protein gcn4 and tropomyosin 1PDBTitle: crystal structure of the c-terminal fragment of rabbit2 skeletal alpha-tropomyosin
54 c2efrB_
not modelled
12.2
9
PDB header: contractile proteinChain: B: PDB Molecule: general control protein gcn4 and tropomyosin 1 alpha chain;PDBTitle: crystal structure of the c-terminal tropomyosin fragment with n- and2 c-terminal extensions of the leucine zipper at 1.8 angstroms3 resolution
55 d2b0ha1
not modelled
12.0
18
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: VBS domain56 c2l7aA_
not modelled
11.6
16
PDB header: cell adhesionChain: A: PDB Molecule: talin-1;PDBTitle: solution structure of the talin vbs2b domain
57 c1wyyB_
not modelled
11.1
15
PDB header: viral proteinChain: B: PDB Molecule: e2 glycoprotein;PDBTitle: post-fusion hairpin conformation of the sars coronavirus spike2 glycoprotein
58 c3arcl_
not modelled
11.0
39
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
59 d1wa8b1
not modelled
10.0
12
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like60 d1i4da_
not modelled
9.2
16
Fold: BAR/IMD domain-likeSuperfamily: BAR/IMD domain-likeFamily: Arfaptin, Rac-binding fragment61 c3arcL_
not modelled
9.0
39
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
62 c3prqL_
not modelled
9.0
39
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
63 c3prrL_
not modelled
9.0
39
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
64 d2axtl1
not modelled
9.0
39
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like65 c1s5lL_
not modelled
9.0
39
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
66 c2axtl_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
67 c2axtL_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
68 c3bz2L_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
69 c3a0hl_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
70 c3a0bl_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
71 c3a0bL_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
72 c3a0hL_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
73 c3kziL_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
74 c1s5ll_
not modelled
9.0
39
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
75 c3bz1L_
not modelled
9.0
39
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
76 c1l4aD_
not modelled
9.0
10
PDB header: endocytosis/exocytosisChain: D: PDB Molecule: s-snap25 fusion protein;PDBTitle: x-ray structure of the neuronal complexin/snare complex2 from the squid loligo pealei
77 c1sfcD_
not modelled
8.7
10
PDB header: transport proteinChain: D: PDB Molecule: protein (snap-25b);PDBTitle: neuronal synaptic fusion complex
78 c3e7kF_
not modelled
8.4
17
PDB header: membrane proteinChain: F: PDB Molecule: trpm7 channel;PDBTitle: crystal structure of an antiparallel coiled-coil tetramerization2 domain from trpm7 channels
79 c2kvpA_
not modelled
8.3
16
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: nmr structure of the talin vbs3 domain, 1815-1973
80 c3dtpA_
not modelled
8.2
15
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
81 d1x79a_
not modelled
8.1
15
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain82 c3gxvD_
not modelled
7.9
23
PDB header: hydrolase/replicationChain: D: PDB Molecule: replicative dna helicase;PDBTitle: three-dimensional structure of n-terminal domain of dnab2 helicase from helicobacter pylori and its interactions with3 primase
83 c2fxmB_
not modelled
7.6
5
PDB header: contractile proteinChain: B: PDB Molecule: myosin heavy chain, cardiac muscle beta isoform;PDBTitle: structure of the human beta-myosin s2 fragment
84 c2npsD_
not modelled
7.6
5
PDB header: transport proteinChain: D: PDB Molecule: syntaxin-6;PDBTitle: crystal structure of the early endosomal snare complex
85 d1o3xa_
not modelled
7.6
14
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain86 c1yvlB_
not modelled
7.5
7
PDB header: signaling proteinChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: structure of unphosphorylated stat1
87 c3gxvC_
not modelled
7.5
23
PDB header: hydrolase/replicationChain: C: PDB Molecule: replicative dna helicase;PDBTitle: three-dimensional structure of n-terminal domain of dnab2 helicase from helicobacter pylori and its interactions with3 primase
88 c1eboE_
not modelled
7.4
18
PDB header: viral proteinChain: E: PDB Molecule: ebola virus envelope protein chimera consistingPDBTitle: crystal structure of the ebola virus membrane-fusion2 subunit, gp2, from the envelope glycoprotein ectodomain
89 d1wrda1
not modelled
7.3
8
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain90 c2j7aC_
not modelled
7.2
24
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c quinol dehydrogenase nrfh;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
91 c2l9uA_
not modelled
7.1
10
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
92 c1junB_
not modelled
7.1
28
PDB header: transcription regulationChain: B: PDB Molecule: c-jun homodimer;PDBTitle: nmr study of c-jun homodimer
93 d1t01a1
not modelled
7.1
12
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin94 d2h8pc1
not modelled
6.8
11
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels95 c2b9cA_
not modelled
6.8
15
PDB header: contractile proteinChain: A: PDB Molecule: striated-muscle alpha tropomyosin;PDBTitle: structure of tropomyosin's mid-region: bending and binding2 sites for actin
96 d1r0da_
not modelled
6.8
15
Fold: I/LWEQ domainSuperfamily: I/LWEQ domainFamily: I/LWEQ domain97 c1m1jA_
not modelled
6.7
5
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha subunit;PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
98 c2l7nA_
not modelled
6.7
9
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: nmr structure of the b domain of talin
99 c1l7cA_
not modelled
6.7
14
PDB header: cell adhesionChain: A: PDB Molecule: alpha e-catenin;PDBTitle: alpha-catenin fragment, residues 385-651