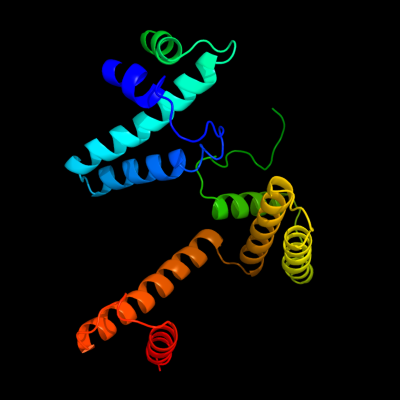
| 1 | c3crcB_
|
|
 |
100.0 |
94 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein mazg;
PDBTitle: crystal structure of escherichia coli mazg, the regulator2 of nutritional stress response
|
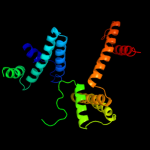
| 2 | c2yxhB_
|
|
 |
100.0 |
26 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:mazg-related protein;
PDBTitle: crystal structure of mazg-related protein from thermotoga maritima
|
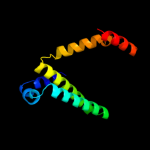
| 3 | d2a3qa1
|
|
 |
99.6 |
17 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
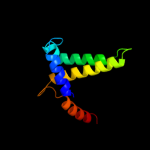
| 4 | c3obcB_
|
|
 |
99.6 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:pyrophosphatase;
PDBTitle: crystal structure of a pyrophosphatase (af1178) from archaeoglobus2 fulgidus at 1.80 a resolution
|
| 5 | c2q4pA_
|
|
 |
99.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein rs21-c6;
PDBTitle: ensemble refinement of the crystal structure of protein from mus2 musculus mm.29898
|
| 6 | d2gtad1
|
|
 |
99.3 |
25 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 7 | d2gtaa1
|
|
 |
99.1 |
27 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 8 | d1vmga_
|
|
 |
99.0 |
20 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 9 | c2q9lA_
|
|
 |
98.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of imazg from vibrio dat 722: ctag-imazg (p43212)
|
| 10 | d2oiea1
|
|
 |
98.3 |
17 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 11 | d1yxba1
|
|
 |
97.5 |
25 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 12 | d1y6xa1
|
|
 |
97.4 |
25 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 13 | c1yvwD_
|
|
 |
97.0 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp2 pyrophosphohydrolase from bacillus cereus. nesgc target3 bcr13.
|
| 14 | d1yvwa1
|
|
 |
97.0 |
18 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 15 | c2a7wF_
|
|
 |
96.9 |
24 |
PDB header:hydrolase
Chain: F: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp pyrophosphatase2 from chromobacterium violaceum (atcc 12472). nesg target3 cvr7
|
| 16 | d2a7wa1
|
|
 |
96.9 |
24 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 17 | c2yf3F_
|
|
 |
93.9 |
32 |
PDB header:hydrolase
Chain: F: PDB Molecule:mazg-like nucleoside triphosphate pyrophosphohydrolase;
PDBTitle: crystal structure of dr2231, the mazg-like protein from2 deinococcus radiodurans, complex with manganese
|
| 18 | c2p06A_
|
|
 |
92.5 |
31 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af_0060;
PDBTitle: crystal structure of a predicted coding region af_0060 from2 archaeoglobus fulgidus dsm 4304
|
| 19 | d2p06a1
|
|
 |
92.5 |
31 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:AF0060-like |
| 20 | c3nqwB_
|
|
 |
68.4 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:cg11900;
PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
|
| 21 | d2f02a1 |
|
not modelled |
56.5 |
22 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 22 | d1zyma1 |
|
not modelled |
53.1 |
11 |
Fold:SAM domain-like
Superfamily:Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain
Family:Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain |
| 23 | c2hroA_ |
|
not modelled |
47.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
| 24 | c3in1A_ |
|
not modelled |
38.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized sugar kinase ydjh;
PDBTitle: crystal structure of a putative ribokinase in complex with2 adp from e.coli
|
| 25 | c2lfcA_ |
|
not modelled |
35.5 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase, flavoprotein subunit;
PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
|
| 26 | c3bf5A_ |
|
not modelled |
35.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:ribokinase related protein;
PDBTitle: crystal structure of putative ribokinase (10640157) from thermoplasma2 acidophilum at 1.91 a resolution
|
| 27 | d2dcna1 |
|
not modelled |
34.0 |
22 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 28 | c2rfpA_ |
|
not modelled |
31.0 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative ntp pyrophosphohydrolase;
PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
|
| 29 | c3ah5E_ |
|
not modelled |
25.7 |
23 |
PDB header:transferase
Chain: E: PDB Molecule:thymidylate synthase thyx;
PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
|
| 30 | d2j4ba1 |
|
not modelled |
24.1 |
19 |
Fold:Taf5 N-terminal domain-like
Superfamily:Taf5 N-terminal domain-like
Family:Taf5 N-terminal domain-like |
| 31 | c2jz8A_ |
|
not modelled |
23.3 |
80 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bh09830;
PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
|
| 32 | c2varB_ |
|
not modelled |
22.5 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:fructokinase;
PDBTitle: crystal structure of sulfolobus solfataricus 2-keto-3-2 deoxygluconate kinase complexed with 2-keto-3-3 deoxygluconate
|
| 33 | d1bpoa1 |
|
not modelled |
21.5 |
60 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Clathrin heavy-chain linker domain |
| 34 | d1qo8a3 |
|
not modelled |
19.7 |
24 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 35 | d2afba1 |
|
not modelled |
19.0 |
15 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 36 | d1y0pa3 |
|
not modelled |
18.8 |
20 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 37 | c3muxB_ |
|
not modelled |
18.4 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:putative 4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
|
| 38 | c2jg5B_ |
|
not modelled |
18.4 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:fructose 1-phosphate kinase;
PDBTitle: crystal structure of a putative phosphofructokinase from2 staphylococcus aureus
|
| 39 | d1vi9a_ |
|
not modelled |
18.1 |
19 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:PfkB-like kinase |
| 40 | c2nwhA_ |
|
not modelled |
18.1 |
17 |
PDB header:signaling protein,transferase
Chain: A: PDB Molecule:carbohydrate kinase;
PDBTitle: carbohydrate kinase from agrobacterium tumefaciens
|
| 41 | d2fv7a1 |
|
not modelled |
18.0 |
30 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 42 | c3m6yA_ |
|
not modelled |
17.9 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
|
| 43 | c3gbuD_ |
|
not modelled |
17.8 |
30 |
PDB header:transferase
Chain: D: PDB Molecule:uncharacterized sugar kinase ph1459;
PDBTitle: crystal structure of an uncharacterized sugar kinase ph1459 from2 pyrococcus horikoshii in complex with atp
|
| 44 | c2jg1C_ |
|
not modelled |
17.7 |
30 |
PDB header:transferase
Chain: C: PDB Molecule:tagatose-6-phosphate kinase;
PDBTitle: structure of staphylococcus aureus d-tagatose-6-phosphate2 kinase with cofactor and substrate
|
| 45 | c3nybA_ |
|
not modelled |
17.6 |
14 |
PDB header:transferase/rna binding protein
Chain: A: PDB Molecule:poly(a) rna polymerase protein 2;
PDBTitle: structure and function of the polymerase core of tramp, a rna2 surveillance complex
|
| 46 | d2fzpa1 |
|
not modelled |
17.4 |
42 |
Fold:NRDP1 C-terminal domain-like
Superfamily:NRDP1 C-terminal domain-like
Family:USP8 interacting domain |
| 47 | d1d4ca3 |
|
not modelled |
17.4 |
20 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 48 | d1omha_ |
|
not modelled |
17.1 |
29 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:Relaxase domain |
| 49 | c2i5bC_ |
|
not modelled |
16.9 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:phosphomethylpyrimidine kinase;
PDBTitle: the crystal structure of an adp complex of bacillus2 subtilis pyridoxal kinase provides evidence for the3 parralel emergence of enzyme activity during evolution
|
| 50 | d3bvua1 |
|
not modelled |
16.7 |
12 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Families 57/38 glycoside transferase middle domain
Family:alpha-mannosidase, domain 2 |
| 51 | d1ogla_ |
|
not modelled |
16.6 |
25 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:Type II deoxyuridine triphosphatase |
| 52 | c3fnnA_ |
|
not modelled |
15.8 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:thymidylate synthase thyx;
PDBTitle: biochemical and structural analysis of an atypical thyx:2 corynebacterium glutamicum nchu 87078 depends on thya for3 thymidine biosynthesis
|
| 53 | d2f2aa1 |
|
not modelled |
15.6 |
19 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
| 54 | c1iclA_ |
|
not modelled |
15.4 |
83 |
PDB header:de novo protein
Chain: A: PDB Molecule:th1ox;
PDBTitle: solution structure of designed beta-sheet mini-protein th1ox
|
| 55 | c3hj6B_ |
|
not modelled |
15.4 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:fructokinase;
PDBTitle: structure of halothermothrix orenii fructokinase (frk)
|
| 56 | c2af6G_ |
|
not modelled |
15.3 |
29 |
PDB header:transferase
Chain: G: PDB Molecule:thymidylate synthase thyx;
PDBTitle: crystal structure of mycobacterium tuberculosis flavin dependent2 thymidylate synthase (mtb thyx) in the presence of co-factor fad and3 substrate analog 5-bromo-2'-deoxyuridine-5'-monophosphate (brdump)
|
| 57 | d1vm7a_ |
|
not modelled |
15.2 |
19 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 58 | d2ibge1 |
|
not modelled |
15.0 |
33 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:Hedgehog (development protein), N-terminal signaling domain |
| 59 | d1yvia1 |
|
not modelled |
14.7 |
2 |
Fold:Four-helical up-and-down bundle
Superfamily:Histidine-containing phosphotransfer domain, HPT domain
Family:Phosphorelay protein-like |
| 60 | d2abqa1 |
|
not modelled |
14.5 |
30 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 61 | c2rbcA_ |
|
not modelled |
14.4 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:sugar kinase;
PDBTitle: crystal structure of a putative ribokinase from agrobacterium2 tumefaciens
|
| 62 | c2ddmA_ |
|
not modelled |
14.2 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxine kinase;
PDBTitle: crystal structure of pyridoxal kinase from the escherichia2 coli pdxk gene at 2.1 a resolution
|
| 63 | c2ld7B_ |
|
not modelled |
14.1 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:paired amphipathic helix protein sin3a;
PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
|
| 64 | c2z36A_ |
|
not modelled |
13.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 type compactin 3'',4''-
PDBTitle: crystal structure of cytochrome p450 moxa from nonomuraea2 recticatena (cyp105)
|
| 65 | c3cqdB_ |
|
not modelled |
13.9 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:6-phosphofructokinase isozyme 2;
PDBTitle: structure of the tetrameric inhibited form of2 phosphofructokinase-2 from escherichia coli
|
| 66 | c3egrB_ |
|
not modelled |
13.9 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phenylacetate-coa oxygenase subunit paab;
PDBTitle: crystal structure of a phenylacetate-coa oxygenase subunit paab2 (reut_a2307) from ralstonia eutropha jmp134 at 2.65 a resolution
|
| 67 | c1bpoA_ |
|
not modelled |
13.8 |
60 |
PDB header:membrane protein
Chain: A: PDB Molecule:protein (clathrin);
PDBTitle: clathrin heavy-chain terminal domain and linker
|
| 68 | c3pl2D_ |
|
not modelled |
13.6 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:sugar kinase, ribokinase family;
PDBTitle: crystal structure of a 5-keto-2-deoxygluconokinase (ncgl0155, cgl0158)2 from corynebacterium glutamicum atcc 13032 kitasato at 1.89 a3 resolution
|
| 69 | d3d1ma1 |
|
not modelled |
13.6 |
33 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:Hedgehog (development protein), N-terminal signaling domain |
| 70 | d1wn0a1 |
|
not modelled |
13.6 |
5 |
Fold:Four-helical up-and-down bundle
Superfamily:Histidine-containing phosphotransfer domain, HPT domain
Family:Phosphorelay protein-like |
| 71 | c3nnqA_ |
|
not modelled |
13.2 |
21 |
PDB header:viral protein
Chain: A: PDB Molecule:n-terminal domain of moloney murine leukemia virus
PDBTitle: crystal structure of the n-terminal domain of moloney murine leukemia2 virus integrase, northeast structural genomics consortium target or3
|
| 72 | c2c49A_ |
|
not modelled |
13.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:sugar kinase mj0406;
PDBTitle: crystal structure of methanocaldococcus jannaschii2 nucleoside kinase - an archaeal member of the ribokinase3 family
|
| 73 | d1ffgb_ |
|
not modelled |
12.9 |
38 |
Fold:Ferredoxin-like
Superfamily:CheY-binding domain of CheA
Family:CheY-binding domain of CheA |
| 74 | c2pmzL_ |
|
not modelled |
12.7 |
18 |
PDB header:translation, transferase
Chain: L: PDB Molecule:dna-directed rna polymerase subunit l;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 75 | c3skqA_ |
|
not modelled |
12.7 |
6 |
PDB header:metal transport
Chain: A: PDB Molecule:mitochondrial distribution and morphology protein 38;
PDBTitle: mdm38 is a 14-3-3-like receptor and associates with the protein2 synthesis machinery at the inner mitochondrial membrane
|
| 76 | c1w7pD_ |
|
not modelled |
12.6 |
32 |
PDB header:protein transport
Chain: D: PDB Molecule:vps36p, ylr417w;
PDBTitle: the crystal structure of endosomal complex escrt-ii2 (vps22/vps25/vps36)
|
| 77 | c1a0oH_ |
|
not modelled |
12.5 |
38 |
PDB header:chemotaxis
Chain: H: PDB Molecule:chea;
PDBTitle: chey-binding domain of chea in complex with chey
|
| 78 | c3lh5A_ |
|
not modelled |
12.5 |
25 |
PDB header:protein binding
Chain: A: PDB Molecule:tight junction protein zo-1;
PDBTitle: crystal structure of the sh3-guanylate kinase core domain of zo-1
|
| 79 | d2ix0a4 |
|
not modelled |
12.4 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:RNB domain-like |
| 80 | c3ebnD_ |
|
not modelled |
12.3 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: a special dimerization of sars-cov main protease c-terminal2 domain due to domain-swapping
|
| 81 | c3m1nB_ |
|
not modelled |
12.1 |
33 |
PDB header:signaling protein
Chain: B: PDB Molecule:sonic hedgehog protein;
PDBTitle: crystal structure of human sonic hedgehog n-terminal domain
|
| 82 | c3e3vA_ |
|
not modelled |
11.9 |
11 |
PDB header:recombination
Chain: A: PDB Molecule:regulatory protein recx;
PDBTitle: crystal structure of recx from lactobacillus salivarius
|
| 83 | d1rkda_ |
|
not modelled |
11.9 |
19 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 84 | c3qk9B_ |
|
not modelled |
11.9 |
20 |
PDB header:protein transport
Chain: B: PDB Molecule:mitochondrial import inner membrane translocase subunit
PDBTitle: yeast tim44 c-terminal domain complexed with cymal-3
|
| 85 | c3kxaD_ |
|
not modelled |
11.4 |
31 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of ngo0477 from neisseria gonorrhoeae
|
| 86 | c3ibqA_ |
|
not modelled |
11.2 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxal kinase;
PDBTitle: crystal structure of pyridoxal kinase from lactobacillus2 plantarum in complex with atp
|
| 87 | c3fymA_ |
|
not modelled |
11.2 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the 1a structure of ymfm, a putative dna-binding membrane2 protein from staphylococcus aureus
|
| 88 | d2absa1 |
|
not modelled |
11.1 |
19 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 89 | c2absA_ |
|
not modelled |
11.1 |
19 |
PDB header:signaling protein,transferase
Chain: A: PDB Molecule:adenosine kinase;
PDBTitle: crystal structure of t. gondii adenosine kinase complexed2 with amp-pcp
|
| 90 | c3kzhA_ |
|
not modelled |
11.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:probable sugar kinase;
PDBTitle: crystal structure of a putative sugar kinase from2 clostridium perfringens
|
| 91 | c3l9qB_ |
|
not modelled |
10.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:dna primase large subunit;
PDBTitle: crystal structure of human polymerase alpha-primase p58 iron-sulfur2 cluster domain
|
| 92 | d1v19a_ |
|
not modelled |
10.8 |
30 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 93 | c3lhxA_ |
|
not modelled |
10.8 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:ketodeoxygluconokinase;
PDBTitle: crystal structure of a ketodeoxygluconokinase (kdgk) from2 shigella flexneri
|
| 94 | d1in0a1 |
|
not modelled |
10.8 |
15 |
Fold:Ferredoxin-like
Superfamily:YajQ-like
Family:YajQ-like |
| 95 | c2dydA_ |
|
not modelled |
10.8 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:poly(a)-binding protein;
PDBTitle: solution structure of the pabc domain from triticum2 aevestium poly(a)-binding protein
|
| 96 | d1bxya_ |
|
not modelled |
10.8 |
24 |
Fold:Ribosomal protein L30p/L7e
Superfamily:Ribosomal protein L30p/L7e
Family:Ribosomal protein L30p/L7e |
| 97 | d1sr2a_ |
|
not modelled |
10.5 |
5 |
Fold:Four-helical up-and-down bundle
Superfamily:Histidine-containing phosphotransfer domain, HPT domain
Family:Sensor-like histidine kinase YojN, C-terminal domain |
| 98 | d1e85a_ |
|
not modelled |
10.5 |
23 |
Fold:Four-helical up-and-down bundle
Superfamily:Cytochromes
Family:Cytochrome c'-like |
| 99 | c2pkkA_ |
|
not modelled |
10.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:adenosine kinase;
PDBTitle: crystal structure of m tuberculosis adenosine kinase complexed with 2-2 fluro adenosine
|