| 1 | d2jo6a1
|
|
 |
100.0 |
100 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 2 | d2jzaa1
|
|
 |
100.0 |
63 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 3 | d3c0da1
|
|
 |
100.0 |
36 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 4 | c2qpzA_
|
|
 |
100.0 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:naphthalene 1,2-dioxygenase system ferredoxin
PDBTitle: naphthalene 1,2-dioxygenase rieske ferredoxin
|
| 5 | d1fqta_
|
|
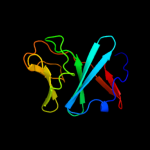 |
100.0 |
21 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 6 | c3gceA_
|
|
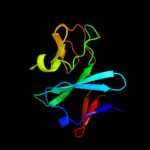 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin component of carbazole 1,9a-
PDBTitle: ferredoxin of carbazole 1,9a-dioxygenase from nocardioides2 aromaticivorans ic177
|
| 7 | c3d89A_
|
|
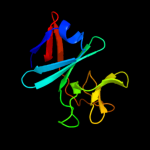 |
99.9 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:rieske domain-containing protein;
PDBTitle: crystal structure of a soluble rieske ferredoxin from mus musculus
|
| 8 | d1z01a1
|
|
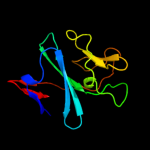 |
99.9 |
13 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 9 | d1vm9a_
|
|
 |
99.9 |
20 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 10 | c2de7E_
|
|
 |
99.9 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ferredoxin component of carbazole;
PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
|
| 11 | d2de6a1
|
|
 |
99.9 |
13 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 12 | c2i7fB_
|
|
 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin component of dioxygenase;
PDBTitle: sphingomonas yanoikuyae b1 ferredoxin
|
| 13 | c2zylA_
|
|
 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:possible oxidoreductase;
PDBTitle: crystal structure of 3-ketosteroid-9-alpha-hydroxylase2 (ksha) from m. tuberculosis
|
| 14 | c3gkqB_
|
|
 |
99.9 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:terminal oxygenase component of carbazole 1,9a-
PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 novosphingobium sp. ka1
|
| 15 | c2de7B_
|
|
 |
99.9 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:terminal oxygenase component of carbazole;
PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
|
| 16 | c1z01D_
|
|
 |
99.9 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:2-oxo-1,2-dihydroquinoline 8-monooxygenase,
PDBTitle: 2-oxoquinoline 8-monooxygenase component: active site2 modulation by rieske-[2fe-2s] center oxidation/reduction
|
| 17 | c3dqyA_
|
|
 |
99.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:toluene 1,2-dioxygenase system ferredoxin
PDBTitle: crystal structure of toluene 2,3-dioxygenase ferredoxin
|
| 18 | c3gcfC_
|
|
 |
99.9 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:terminal oxygenase component of carbazole 1,9a-
PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 nocardioides aromaticivorans ic177
|
| 19 | c3gteB_
|
|
 |
99.9 |
20 |
PDB header:electron transport, oxidoreductase
Chain: B: PDB Molecule:ddmc;
PDBTitle: crystal structure of dicamba monooxygenase with non-heme2 iron
|
| 20 | c3n0qA_
|
|
 |
99.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative aromatic-ring hydroxylating dioxygenase;
PDBTitle: crystal structure of a putative aromatic-ring hydroxylating2 dioxygenase (tm1040_3219) from silicibacter sp. tm1040 at 1.80 a3 resolution
|
| 21 | d1ulia1 |
|
not modelled |
99.9 |
12 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 22 | d1wqla1 |
|
not modelled |
99.9 |
11 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 23 | d1o7na1 |
|
not modelled |
99.9 |
19 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 24 | d2bmoa1 |
|
not modelled |
99.9 |
21 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 25 | c1uljA_ |
|
not modelled |
99.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biphenyl dioxygenase large subunit;
PDBTitle: biphenyl dioxygenase (bpha1a2) in complex with the substrate
|
| 26 | c1wqlA_ |
|
not modelled |
99.9 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-sulfur protein large subunit of cumene dioxygenase;
PDBTitle: cumene dioxygenase (cuma1a2) from pseudomonas fluorescens ip01
|
| 27 | d2b1xa1 |
|
not modelled |
99.9 |
11 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 28 | c2hmnA_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:naphthalene 1,2-dioxygenase alpha subunit;
PDBTitle: crystal structure of the naphthalene 1,2-dioxygenase f352v2 mutant bound to anthracene.
|
| 29 | c2b1xE_ |
|
not modelled |
99.9 |
12 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:naphthalene dioxygenase large subunit;
PDBTitle: crystal structure of naphthalene 1,2-dioxygenase from rhodococcus sp.
|
| 30 | c2gbxE_ |
|
not modelled |
99.9 |
12 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:biphenyl 2,3-dioxygenase alpha subunit;
PDBTitle: crystal structure of biphenyl 2,3-dioxygenase from sphingomonas2 yanoikuyae b1 bound to biphenyl
|
| 31 | d1g8kb_ |
|
not modelled |
99.8 |
10 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 32 | d1rfsa_ |
|
not modelled |
99.8 |
13 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 33 | d3cx5e1 |
|
not modelled |
99.8 |
13 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 34 | d1q90c_ |
|
not modelled |
99.8 |
11 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 35 | d1nyka_ |
|
not modelled |
99.7 |
20 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 36 | d2e74d1 |
|
not modelled |
99.7 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 37 | c2e76D_ |
|
not modelled |
99.7 |
15 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
| 38 | d1riea_ |
|
not modelled |
99.6 |
10 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 39 | c1p84E_ |
|
not modelled |
99.6 |
11 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
| 40 | c2fyuE_ |
|
not modelled |
99.5 |
10 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
| 41 | c2fynO_ |
|
not modelled |
99.5 |
16 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
| 42 | c2nvgA_ |
|
not modelled |
99.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: soluble domain of rieske iron sulfur protein.
|
| 43 | d1jm1a_ |
|
not modelled |
99.3 |
16 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 44 | d2hf1a1 |
|
not modelled |
58.6 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
| 45 | d2jnya1 |
|
not modelled |
56.6 |
22 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
| 46 | d2pk7a1 |
|
not modelled |
52.9 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
| 47 | c2jr6A_ |
|
not modelled |
49.4 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0434 protein nma0874;
PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
|
| 48 | c2kpiA_ |
|
not modelled |
47.9 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sco3027;
PDBTitle: solution nmr structure of streptomyces coelicolor sco30272 modeled with zn+2 bound, northeast structural genomics3 consortium target rr58
|
| 49 | c2js4A_ |
|
not modelled |
45.6 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0434 protein bb2007;
PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
|
| 50 | c3lasA_ |
|
not modelled |
31.5 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:putative carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
|
| 51 | c2w3nA_ |
|
not modelled |
27.5 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase 2;
PDBTitle: structure and inhibition of the co2-sensing carbonic2 anhydrase can2 from the pathogenic fungus cryptococcus3 neoformans
|
| 52 | c1ylkA_ |
|
not modelled |
27.4 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein rv1284/mt1322;
PDBTitle: crystal structure of rv1284 from mycobacterium tuberculosis in complex2 with thiocyanate
|
| 53 | d1ddza1 |
|
not modelled |
26.9 |
17 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 54 | c2kppA_ |
|
not modelled |
26.3 |
3 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin0431 protein;
PDBTitle: solution nmr structure of lin0431 protein from listeria innocua.2 northeast structural genomics consortium target lkr112
|
| 55 | c2a8cE_ |
|
not modelled |
26.2 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:carbonic anhydrase 2;
PDBTitle: haemophilus influenzae beta-carbonic anhydrase
|
| 56 | d1ddza2 |
|
not modelled |
26.2 |
17 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 57 | c1ddzA_ |
|
not modelled |
26.2 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: x-ray structure of a beta-carbonic anhydrase from the red2 alga, porphyridium purpureum r-1
|
| 58 | c3eyxB_ |
|
not modelled |
25.2 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
|
| 59 | d1g5ca_ |
|
not modelled |
22.0 |
8 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 60 | d2aqaa1 |
|
not modelled |
22.0 |
17 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 61 | c3ucoB_ |
|
not modelled |
21.6 |
18 |
PDB header:lyase/lyase inhibitor
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: coccomyxa beta-carbonic anhydrase in complex with iodide
|
| 62 | c2a5vB_ |
|
not modelled |
21.6 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase (carbonate dehydratase) (carbonic
PDBTitle: crystal structure of m. tuberculosis beta carbonic anhydrase, rv3588c,2 tetrameric form
|
| 63 | d1i6pa_ |
|
not modelled |
21.5 |
27 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 64 | d1ekja_ |
|
not modelled |
21.2 |
17 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 65 | d1t1ua1 |
|
not modelled |
17.9 |
15 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:Choline/Carnitine O-acyltransferase |
| 66 | c1t7qA_ |
|
not modelled |
13.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:carnitine acetyltransferase;
PDBTitle: crystal structure of the f565a mutant of murine carnitine2 acetyltransferase in complex with carnitine and coa
|
| 67 | c2h4tB_ |
|
not modelled |
12.3 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:carnitine o-palmitoyltransferase ii,
PDBTitle: crystal structure of rat carnitine palmitoyltransferase ii
|
| 68 | d1xl7a1 |
|
not modelled |
12.2 |
13 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:Choline/Carnitine O-acyltransferase |
| 69 | c2j6aA_ |
|
not modelled |
11.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:protein trm112;
PDBTitle: crystal structure of s. cerevisiae ynr046w, a zinc finger2 protein from the erf1 methyltransferase complex.
|
| 70 | c1q6xA_ |
|
not modelled |
11.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:choline o-acetyltransferase;
PDBTitle: crystal structure of rat choline acetyltransferase
|
| 71 | d1ubdc1 |
|
not modelled |
11.4 |
18 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 72 | d1ndba1 |
|
not modelled |
11.3 |
15 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:Choline/Carnitine O-acyltransferase |
| 73 | d2j7ja2 |
|
not modelled |
10.5 |
17 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 74 | c2jyeA_ |
|
not modelled |
10.4 |
16 |
PDB header:cytokine
Chain: A: PDB Molecule:granulin a;
PDBTitle: human granulin a
|
| 75 | d2apob1 |
|
not modelled |
9.9 |
29 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 76 | d1nm8a1 |
|
not modelled |
9.5 |
18 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:Choline/Carnitine O-acyltransferase |
| 77 | d2bdra1 |
|
not modelled |
9.3 |
9 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Ureidoglycolate hydrolase AllA |
| 78 | c2fyoA_ |
|
not modelled |
9.1 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:carnitine o-palmitoyltransferase ii,
PDBTitle: crystal structure of rat carnitine palmitoyltransferase 22 in space group p43212
|
| 79 | d2nn6g3 |
|
not modelled |
8.9 |
13 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 80 | d2c42a2 |
|
not modelled |
8.8 |
8 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:PFOR PP module |
| 81 | c2fy2A_ |
|
not modelled |
8.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:choline o-acetyltransferase;
PDBTitle: structures of ligand bound human choline acetyltransferase2 provide insight into regulation of acetylcholine synthesis
|
| 82 | d2ey4e1 |
|
not modelled |
8.7 |
21 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 83 | c3izbP_ |
|
not modelled |
8.1 |
20 |
PDB header:ribosome
Chain: P: PDB Molecule:40s ribosomal protein rps11 (s17p);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 84 | c2k5rA_ |
|
not modelled |
7.9 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein xf2673;
PDBTitle: solution nmr structure of xf2673 from xylella fastidiosa.2 northeast structural genomics consortium target xfr39
|
| 85 | c3iz6P_ |
|
not modelled |
7.8 |
20 |
PDB header:ribosome
Chain: P: PDB Molecule:40s ribosomal protein s11 (s17p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 86 | d1l1oc_ |
|
not modelled |
7.6 |
9 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 87 | c2xznQ_ |
|
not modelled |
6.7 |
20 |
PDB header:ribosome
Chain: Q: PDB Molecule:ribosomal protein s17 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 88 | d1xsqa_ |
|
not modelled |
6.3 |
17 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Ureidoglycolate hydrolase AllA |
| 89 | d2ct7a1 |
|
not modelled |
6.2 |
15 |
Fold:RING/U-box
Superfamily:RING/U-box
Family:IBR domain |
| 90 | c2aklA_ |
|
not modelled |
6.1 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phna-like protein pa0128;
PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
|
| 91 | d1hyoa1 |
|
not modelled |
5.9 |
13 |
Fold:SH3-like barrel
Superfamily:Fumarylacetoacetate hydrolase, FAH, N-terminal domain
Family:Fumarylacetoacetate hydrolase, FAH, N-terminal domain |
| 92 | c2c3yA_ |
|
not modelled |
5.8 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate-ferredoxin oxidoreductase;
PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
|
| 93 | d2ja9a2 |
|
not modelled |
5.7 |
13 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 94 | c1skzA_ |
|
not modelled |
5.5 |
9 |
PDB header:serine protease inhibitor
Chain: A: PDB Molecule:antistasin;
PDBTitle: protease inhibitor
|
| 95 | d1mdah_ |
|
not modelled |
5.3 |
14 |
Fold:7-bladed beta-propeller
Superfamily:YVTN repeat-like/Quinoprotein amine dehydrogenase
Family:Methylamine dehydrogenase, H-chain |
| 96 | d1gpca_ |
|
not modelled |
5.2 |
27 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Phage ssDNA-binding proteins |