| 1 | c1z1bA_
|
|
 |
100.0 |
33 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:integrase;
PDBTitle: crystal structure of a lambda integrase dimer bound to a2 coc' core site
|
| 2 | d1p7da_
|
|
 |
100.0 |
36 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 3 | c1ma7A_
|
|
 |
100.0 |
12 |
PDB header:hydrolase, ligase/dna
Chain: A: PDB Molecule:cre recombinase;
PDBTitle: crystal structure of cre site-specific recombinase2 complexed with a mutant dna substrate, loxp-a8/t27
|
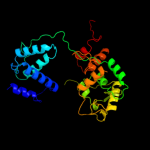
| 4 | c1crxA_
|
|
 |
100.0 |
11 |
PDB header:replication/dna
Chain: A: PDB Molecule:cre recombinase;
PDBTitle: cre recombinase/dna complex reaction intermediate i
|
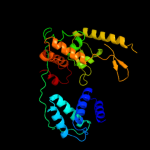
| 5 | c2a3vA_
|
|
 |
100.0 |
17 |
PDB header:recombination
Chain: A: PDB Molecule:site-specific recombinase inti4;
PDBTitle: structural basis for broad dna-specificity in integron2 recombination
|
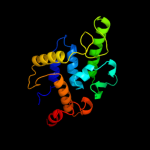
| 6 | c3nkhB_
|
|
 |
100.0 |
17 |
PDB header:recombination
Chain: B: PDB Molecule:integrase;
PDBTitle: crystal structure of integrase from mrsa strain staphylococcus aureus
|
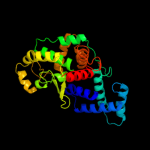
| 7 | c1a0pA_
|
|
 |
100.0 |
16 |
PDB header:dna recombination
Chain: A: PDB Molecule:site-specific recombinase xerd;
PDBTitle: site-specific recombinase, xerd
|
| 8 | d1ae9a_
|
|
 |
99.9 |
36 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 9 | d1aiha_
|
|
 |
99.9 |
17 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 10 | d1f44a2
|
|
 |
99.9 |
13 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 11 | d1a0pa2
|
|
 |
99.9 |
17 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 12 | d5crxb2
|
|
 |
99.8 |
16 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 13 | c2khqA_
|
|
 |
99.1 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution nmr structure of a phage integrase ssp19472 fragment 59-159 from staphylococcus saprophyticus,3 northeast structural genomics consortium target syr103b
|
| 14 | c2oxoA_
|
|
 |
99.1 |
28 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: crystallization and structure determination of the core-2 binding domain of bacteriophage lambda integrase
|
| 15 | c2kiwA_
|
|
 |
99.0 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:int protein;
PDBTitle: solution nmr structure of the domain n-terminal to the2 integrase domain of sh1003 from staphylococcus3 haemolyticus. northeast structural genomics consortium4 target shr105f (64-166).
|
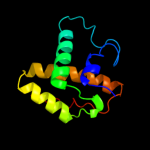
| 16 | c3lysC_
|
|
 |
99.0 |
13 |
PDB header:recombination
Chain: C: PDB Molecule:prophage pi2 protein 01, integrase;
PDBTitle: crystal structure of the n-terminal domain of the prophage2 pi2 protein 01 (integrase) from lactococcus lactis,3 northeast structural genomics consortium target kr124f
|
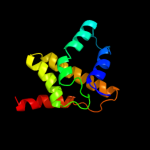
| 17 | c2kj8A_
|
|
 |
98.8 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative prophage cps-53 integrase;
PDBTitle: nmr structure of fragment 87-196 from the putative phage2 integrase ints of e. coli: northeast structural genomics3 consortium target er652a, psi-2
|
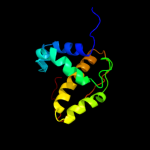
| 18 | c2kkvA_
|
|
 |
98.8 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution nmr structure of an integrase domain from protein2 spa4288 from salmonella enterica, northeast structural3 genomics consortium target slr105h
|
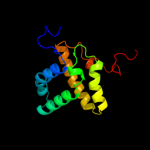
| 19 | c2kj9A_
|
|
 |
98.8 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: nmr structure of intb phage-integrase-like protein fragment2 90-199 from erwinia carotova subsp. atroseptica: northeast3 structural genomics consortium target ewr217e
|
| 20 | c2kobA_
|
|
 |
98.7 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of clolep_01837 (fragment 61-160)2 from clostridium leptum. northeast structural genomics3 consortium target qlr8a
|
| 21 | c2kd1A_ |
|
not modelled |
98.6 |
5 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dna integration/recombination/invertion protein;
PDBTitle: solution nmr structure of the integrase-like domain from2 bacillus cereus ordered locus bc_1272. northeast3 structural genomics consortium target bcr268f
|
| 22 | c2kkpA_ |
|
not modelled |
98.6 |
16 |
PDB header:dna binding protein
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of the phage integrase sam-like2 domain from moth 1796 from moorella thermoacetica.3 northeast structural genomics consortium target mtr39k4 (residues 64-171).
|
| 23 | c2khvA_ |
|
not modelled |
98.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of protein nmul_a0922 from2 nitrosospira multiformis. northeast structural genomics3 consortium target nmr38b.
|
| 24 | c2kj5A_ |
|
not modelled |
98.5 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of a domain from a putative phage2 integrase protein nmul_a0064 from nitrosospira multiformis,3 northeast structural genomics consortium target nmr46c
|
| 25 | c2keyA_ |
|
not modelled |
98.4 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative phage integrase;
PDBTitle: solution nmr structure of a domain from a putative phage integrase2 protein bf2284 from bacteroides fragilis, northeast structural3 genomics consortium target bfr257c
|
| 26 | c2v6eB_ |
|
not modelled |
98.2 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:protelemorase;
PDBTitle: protelomerase telk complexed with substrate dna
|
| 27 | d1z1ba1 |
|
not modelled |
98.1 |
41 |
Fold:DNA-binding domain
Superfamily:DNA-binding domain
Family:lambda integrase N-terminal domain |
| 28 | c3nrwA_ |
|
not modelled |
98.1 |
12 |
PDB header:recombination
Chain: A: PDB Molecule:phage integrase/site-specific recombinase;
PDBTitle: crystal structure of the n-terminal domain of phage integrase/site-2 specific recombinase (tnp) from haloarcula marismortui, northeast3 structural genomics consortium target hmr208a
|
| 29 | c1kjkA_ |
|
not modelled |
98.1 |
41 |
PDB header:viral protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution structure of the lambda integrase amino-terminal2 domain
|
| 30 | c3ju0A_ |
|
not modelled |
97.9 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:phage integrase;
PDBTitle: structure of the arm-type binding domain of hai7 integrase
|
| 31 | c3jtzA_ |
|
not modelled |
97.5 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: structure of the arm-type binding domain of hpi integrase
|
| 32 | d1a0pa1 |
|
not modelled |
96.9 |
14 |
Fold:SAM domain-like
Superfamily:lambda integrase-like, N-terminal domain
Family:lambda integrase-like, N-terminal domain |
| 33 | d1f44a1 |
|
not modelled |
95.0 |
12 |
Fold:SAM domain-like
Superfamily:lambda integrase-like, N-terminal domain
Family:lambda integrase-like, N-terminal domain |
| 34 | c2b9sA_ |
|
not modelled |
77.9 |
16 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:topoisomerase i-like protein;
PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
|
| 35 | c1a31A_ |
|
not modelled |
77.2 |
16 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:protein (topoisomerase i);
PDBTitle: human reconstituted dna topoisomerase i in covalent complex2 with a 22 base pair dna duplex
|
| 36 | d1p4ea2 |
|
not modelled |
74.6 |
17 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 37 | d1k4ta2 |
|
not modelled |
74.2 |
14 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 38 | c1p4eB_ |
|
not modelled |
72.8 |
17 |
PDB header:dna binding protein/recombination/dna
Chain: B: PDB Molecule:recombinase flp protein;
PDBTitle: flpe w330f mutant-dna holliday junction complex
|
| 39 | d1rr8c1 |
|
not modelled |
69.9 |
14 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 40 | c2f4qA_ |
|
not modelled |
47.3 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:type i topoisomerase, putative;
PDBTitle: crystal structure of deinococcus radiodurans topoisomerase ib
|
| 41 | c1nh3A_ |
|
not modelled |
35.5 |
16 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:dna topoisomerase i;
PDBTitle: human topoisomerase i ara-c complex
|
| 42 | c2h7fX_ |
|
not modelled |
28.3 |
15 |
PDB header:isomerase/dna
Chain: X: PDB Molecule:dna topoisomerase 1;
PDBTitle: structure of variola topoisomerase covalently bound to dna
|
| 43 | c2k49A_ |
|
not modelled |
14.5 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0339 protein so_3888;
PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
|
| 44 | d2csba5 |
|
not modelled |
14.4 |
27 |
Fold:Topoisomerase V catalytic domain-like
Superfamily:Topoisomerase V catalytic domain-like
Family:Topoisomerase V catalytic domain-like |
| 45 | c3m6zA_ |
|
not modelled |
12.0 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:topoisomerase v;
PDBTitle: crystal structure of an n-terminal 44 kda fragment of topoisomerase v2 in the presence of guanidium hydrochloride
|
| 46 | c3igmA_ |
|
not modelled |
11.4 |
6 |
PDB header:transcription/dna
Chain: A: PDB Molecule:pf14_0633 protein;
PDBTitle: a 2.2a crystal structure of the ap2 domain of pf14_0633 from p.2 falciparum, bound as a domain-swapped dimer to its cognate dna
|
| 47 | d1tc3c_ |
|
not modelled |
10.1 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 48 | c2k8eA_ |
|
not modelled |
10.0 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0339 protein yegp;
PDBTitle: solution nmr structure of protein of unknown function yegp from e.2 coli. ontario center for structural proteomics target ec0640_1_1233 northeast structural genomics consortium target et102.
|
| 49 | c2csdB_ |
|
not modelled |
10.0 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:topoisomerase v;
PDBTitle: crystal structure of topoisomerase v (61 kda fragment)
|
| 50 | d2k49a2 |
|
not modelled |
7.8 |
11 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 51 | d2k8ea1 |
|
not modelled |
7.5 |
11 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 52 | c2rrhA_ |
|
not modelled |
7.4 |
30 |
PDB header:hormone
Chain: A: PDB Molecule:vip peptides;
PDBTitle: nmr structure of vasoactive intestinal peptide in methanol
|
| 53 | d1o66a_ |
|
not modelled |
7.1 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 54 | c2ka4B_ |
|
not modelled |
6.4 |
33 |
PDB header:transcription regulator
Chain: B: PDB Molecule:signal transducer and activator of transcription
PDBTitle: nmr structure of the cbp-taz1/stat2-tad complex
|
| 55 | c2d2pA_ |
|
not modelled |
6.4 |
30 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:pituitary adenylate cyclase activating
PDBTitle: the solution structure of micelle-bound peptide
|
| 56 | c1yd6A_ |
|
not modelled |
6.2 |
16 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uvrc;
PDBTitle: crystal structure of the giy-yig n-terminal endonuclease2 domain of uvrc from bacillus caldotenax
|
| 57 | d2al3a1 |
|
not modelled |
5.9 |
9 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:UBX domain |
| 58 | c2kvvA_ |
|
not modelled |
5.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative excisionase;
PDBTitle: solution nmr of putative excisionase from klebsiella pneumoniae,2 northeast structural genomics consortium target target kpr49
|
| 59 | d1ug2a_ |
|
not modelled |
5.4 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 60 | d1j6xa_ |
|
not modelled |
5.3 |
16 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 61 | d2bgxa2 |
|
not modelled |
5.2 |
8 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 62 | d1d1da2 |
|
not modelled |
5.1 |
18 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 63 | c2kgfA_ |
|
not modelled |
5.1 |
12 |
PDB header:viral protein
Chain: A: PDB Molecule:capsid protein p27;
PDBTitle: n-terminal domain of capsid protein from the mason-pfizer2 monkey virus
|
| 64 | d1j2oa2 |
|
not modelled |
5.1 |
44 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |