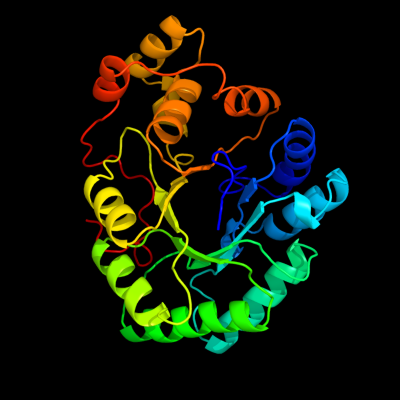
| 1 | c3up8B_
|
|
 |
100.0 |
44 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative 2,5-diketo-d-gluconic acid reductase b;
PDBTitle: crystal structure of a putative 2,5-diketo-d-gluconic acid reductase b
|
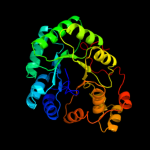
| 2 | d1q5ma_
|
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
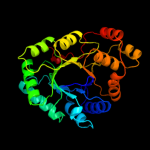
| 3 | d1frba_
|
|
 |
100.0 |
34 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
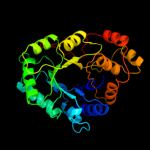
| 4 | d1hqta_
|
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 5 | c3h7uA_
|
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c9
|
| 6 | d1j96a_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 7 | c3f7jB_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:yvgn protein;
PDBTitle: b.subtilis yvgn
|
| 8 | d2alra_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 9 | d1mi3a_
|
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 10 | d1ah4a_
|
|
 |
100.0 |
36 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 11 | d1afsa_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 12 | d1qwka_
|
|
 |
100.0 |
36 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 13 | d1c9wa_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 14 | d1us0a_
|
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 15 | d1s1pa_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 16 | c2bgsA_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldose reductase;
PDBTitle: holo aldose reductase from barley
|
| 17 | c2wztA_
|
|
 |
100.0 |
37 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of a mycobacterium aldo-keto reductase in2 its apo and liganded form
|
| 18 | d1mzra_
|
|
 |
100.0 |
38 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 19 | c3buvB_
|
|
 |
100.0 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxo-5-beta-steroid 4-dehydrogenase;
PDBTitle: crystal structure of human delta(4)-3-ketosteroid 5-beta-reductase in2 complex with nadp and hepes. resolution: 1.35 a.
|
| 20 | c1zgdB_
|
|
 |
100.0 |
32 |
PDB header:plant protein
Chain: B: PDB Molecule:chalcone reductase;
PDBTitle: chalcone reductase complexed with nadp+ at 1.7 angstrom2 resolution
|
| 21 | c3h7rA_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c8
|
| 22 | c3b3dA_ |
|
not modelled |
100.0 |
37 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative morphine dehydrogenase;
PDBTitle: b.subtilis ytbe
|
| 23 | d1vp5a_ |
|
not modelled |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 24 | c1vbjB_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin f synthase;
PDBTitle: the crystal structure of prostaglandin f synthase from2 trypanosoma brucei
|
| 25 | c3o0kB_ |
|
not modelled |
100.0 |
37 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aldo/keto reductase;
PDBTitle: crystal structure of aldo/keto reductase from brucella melitensis
|
| 26 | d1lqaa_ |
|
not modelled |
100.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 27 | d1hw6a_ |
|
not modelled |
100.0 |
42 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 28 | c3krbB_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aldose reductase;
PDBTitle: structure of aldose reductase from giardia lamblia at 1.75a resolution
|
| 29 | d3eaua1 |
|
not modelled |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 30 | c3lutA_ |
|
not modelled |
100.0 |
27 |
PDB header:membrane protein
Chain: A: PDB Molecule:voltage-gated potassium channel subunit beta-2;
PDBTitle: a structural model for the full-length shaker potassium channel kv1.2
|
| 31 | d1ur3m_ |
|
not modelled |
100.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 32 | c3erpA_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of idp01002, a putative oxidoreductase from and essential2 gene of salmonella typhimurium
|
| 33 | d1gvea_ |
|
not modelled |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 34 | c3n6qF_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:yghz aldo-keto reductase;
PDBTitle: crystal structure of yghz from e. coli
|
| 35 | d1pyfa_ |
|
not modelled |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 36 | c3n2tA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the glycerol dehydrogenase akr11b4 from gluconobacter2 oxydans
|
| 37 | d1pz1a_ |
|
not modelled |
100.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 38 | c2bp1C_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:aflatoxin b1 aldehyde reductase member 2;
PDBTitle: structure of the aflatoxin aldehyde reductase in complex2 with nadph
|
| 39 | c1ynpA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: aldo-keto reductase akr11c1 from bacillus halodurans (apo form)
|
| 40 | c3ln3A_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodiol dehydrogenase;
PDBTitle: crystal structure of putative reductase (np_038806.2) from2 mus musculus at 1.18 a resolution
|
| 41 | c3g8rA_ |
|
not modelled |
82.1 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
| 42 | c3ngfA_ |
|
not modelled |
79.8 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 43 | c1xuzA_ |
|
not modelled |
68.3 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 44 | d1jpma1 |
|
not modelled |
58.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 45 | d2zdra2 |
|
not modelled |
52.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 46 | d1k77a_ |
|
not modelled |
44.0 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Hypothetical protein YgbM (EC1530) |
| 47 | d1gqna_ |
|
not modelled |
40.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 48 | c1vliA_ |
|
not modelled |
37.7 |
8 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
| 49 | d7odca2 |
|
not modelled |
35.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 50 | c2ph5A_ |
|
not modelled |
34.5 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
| 51 | d2ijqa1 |
|
not modelled |
33.5 |
12 |
Fold:Hyaluronidase domain-like
Superfamily:TTHA0068-like
Family:TTHA0068-like |
| 52 | d2q02a1 |
|
not modelled |
32.3 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 53 | d1vp8a_ |
|
not modelled |
29.1 |
18 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 54 | c3js3C_ |
|
not modelled |
29.0 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 55 | c3l2iB_ |
|
not modelled |
28.6 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 56 | c3aamA_ |
|
not modelled |
27.7 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease iv;
PDBTitle: crystal structure of endonuclease iv from thermus thermophilus hb8
|
| 57 | d3bofa2 |
|
not modelled |
27.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 58 | d1i27a_ |
|
not modelled |
26.4 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal domain of the rap74 subunit of TFIIF |
| 59 | d1vlia2 |
|
not modelled |
23.3 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 60 | c3cnyA_ |
|
not modelled |
22.0 |
10 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
| 61 | c3oa3A_ |
|
not modelled |
20.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 coccidioides immitis
|
| 62 | c3smaD_ |
|
not modelled |
20.3 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:frbf;
PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
|
| 63 | d2plca_ |
|
not modelled |
20.0 |
46 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Bacterial PLC |
| 64 | d1j5ya2 |
|
not modelled |
20.0 |
15 |
Fold:HPr-like
Superfamily:Putative transcriptional regulator TM1602, C-terminal domain
Family:Putative transcriptional regulator TM1602, C-terminal domain |
| 65 | d1r0ma1 |
|
not modelled |
19.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 66 | c3ic5A_ |
|
not modelled |
19.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 67 | d1xp3a1 |
|
not modelled |
19.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 68 | d1nu5a1 |
|
not modelled |
18.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 69 | c3dx5A_ |
|
not modelled |
18.4 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 70 | c3ktcB_ |
|
not modelled |
17.8 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
| 71 | d2ptda_ |
|
not modelled |
16.5 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Bacterial PLC |
| 72 | c2p0oA_ |
|
not modelled |
16.2 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf871;
PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
|
| 73 | d2gdqa1 |
|
not modelled |
16.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 74 | c3gpqA_ |
|
not modelled |
15.6 |
22 |
PDB header:viral protein/rna
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of macro domain of chikungunya virus in complex with2 rna
|
| 75 | d1fuia2 |
|
not modelled |
15.2 |
14 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
| 76 | d1x87a_ |
|
not modelled |
15.0 |
15 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |
| 77 | d1ub3a_ |
|
not modelled |
14.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 78 | d1o0ya_ |
|
not modelled |
14.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 79 | d1pv8a_ |
|
not modelled |
14.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 80 | c2zvrA_ |
|
not modelled |
13.6 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
|
| 81 | c3kwsB_ |
|
not modelled |
13.5 |
7 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
| 82 | c3e4fB_ |
|
not modelled |
12.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:aminoglycoside n3-acetyltransferase;
PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
|
| 83 | d1u83a_ |
|
not modelled |
12.4 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
| 84 | c1u83A_ |
|
not modelled |
12.4 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:phosphosulfolactate synthase;
PDBTitle: psl synthase from bacillus subtilis
|
| 85 | c2cg8B_ |
|
not modelled |
11.7 |
15 |
PDB header:lyase/transferase
Chain: B: PDB Molecule:dihydroneopterin aldolase 6-hydroxymethyl-7,8-
PDBTitle: the bifunctional dihydroneopterin aldolase 6-hydroxymethyl-2 7,8-dihydropterin synthase from streptococcus pneumoniae
|
| 86 | c3dnfB_ |
|
not modelled |
10.9 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
| 87 | d1ulza2 |
|
not modelled |
10.9 |
31 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 88 | d1sjda1 |
|
not modelled |
10.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 89 | c3gqeA_ |
|
not modelled |
10.6 |
30 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of macro domain of venezuelan equine encephalitis2 virus
|
| 90 | d1spva_ |
|
not modelled |
10.2 |
14 |
Fold:Macro domain-like
Superfamily:Macro domain-like
Family:Macro domain |
| 91 | d1nexa1 |
|
not modelled |
10.1 |
17 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 92 | c1x7fA_ |
|
not modelled |
10.1 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:outer surface protein;
PDBTitle: crystal structure of an uncharacterized b. cereus protein
|
| 93 | d2cp6a1 |
|
not modelled |
9.9 |
23 |
Fold:SH3-like barrel
Superfamily:Cap-Gly domain
Family:Cap-Gly domain |
| 94 | c2fknC_ |
|
not modelled |
9.5 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:urocanate hydratase;
PDBTitle: crystal structure of urocanase from bacillus subtilis
|
| 95 | d2jn6a1 |
|
not modelled |
9.4 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Cgl2762-like |
| 96 | d2chra1 |
|
not modelled |
9.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 97 | c2nx9B_ |
|
not modelled |
9.3 |
32 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 98 | d1geqa_ |
|
not modelled |
9.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 99 | d1uwka_ |
|
not modelled |
9.2 |
13 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |