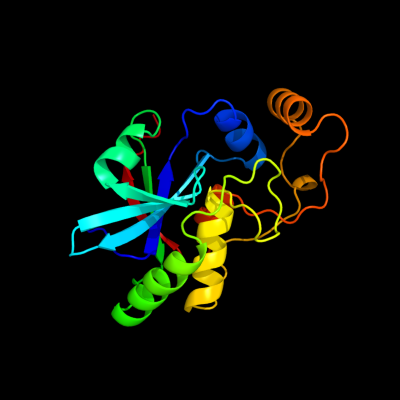
| 1 | d2if6a1
|
|
 |
100.0 |
98 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:YiiX-like |
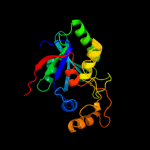
| 2 | c3kw0D_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:cysteine peptidase;
PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
|
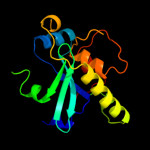
| 3 | c2p1gA_
|
|
 |
98.1 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative xylanase;
PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
|
| 4 | c2k1gA_
|
|
 |
97.8 |
15 |
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein spr;
PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
|
| 5 | c3npfB_
|
|
 |
97.8 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative dipeptidyl-peptidase vi;
PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
|
| 6 | c2fg0B_
|
|
 |
97.6 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 7 | c3gt2A_
|
|
 |
97.5 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 8 | c3h41A_
|
|
 |
97.5 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60 family protein;
PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
|
| 9 | d2evra2
|
|
 |
97.5 |
20 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 10 | c2kytA_
|
|
 |
97.5 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:group xvi phospholipase a2;
PDBTitle: solution struture of the h-rev107 n-terminal domain
|
| 11 | c2xivA_
|
|
 |
97.2 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 12 | c3pbiA_
|
|
 |
97.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 13 | c3i86A_
|
|
 |
97.2 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
|
| 14 | d2im9a1
|
|
 |
97.1 |
20 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Lpg0564-like |
| 15 | c2im9A_
|
|
 |
97.1 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of protein lpg0564 from legionella pneumophila str.2 philadelphia 1, pfam duf1460
|
| 16 | c3m1uB_
|
|
 |
94.3 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative gamma-d-glutamyl-l-diamino acid endopeptidase;
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
|
| 17 | d2io8a2
|
|
 |
92.8 |
16 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:CHAP domain |
| 18 | c2ioaA_
|
|
 |
80.2 |
17 |
PDB header:ligase, hydrolase
Chain: A: PDB Molecule:bifunctional glutathionylspermidine
PDBTitle: e. coli bifunctional glutathionylspermidine2 synthetase/amidase incomplex with mg2+ and adp and3 phosphinate inhibitor
|
| 19 | c2k3aA_
|
|
 |
77.4 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:chap domain protein;
PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
|
| 20 | c2vpmB_
|
|
 |
77.2 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:trypanothione synthetase;
PDBTitle: trypanothione synthetase
|
| 21 | d1u5tb1 |
|
not modelled |
33.0 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 22 | d1wh7a_ |
|
not modelled |
23.5 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 23 | c3mvnA_ |
|
not modelled |
23.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
| 24 | d1wfza_ |
|
not modelled |
20.6 |
19 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 25 | d1zj8a1 |
|
not modelled |
19.9 |
27 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 26 | c1wypA_ |
|
not modelled |
18.7 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:calponin 1;
PDBTitle: solution structure of the ch domain of human calponin 1
|
| 27 | c2z7eB_ |
|
not modelled |
17.9 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:nifu-like protein;
PDBTitle: crystal structure of aquifex aeolicus iscu with bound [2fe-2 2s] cluster
|
| 28 | d1jala2 |
|
not modelled |
13.6 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:TGS-like
Family:G domain-linked domain |
| 29 | d1vbga2 |
|
not modelled |
13.3 |
18 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 30 | d1ni3a2 |
|
not modelled |
13.1 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:TGS-like
Family:G domain-linked domain |
| 31 | d1knwa1 |
|
not modelled |
12.9 |
7 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 32 | d1vcta2 |
|
not modelled |
12.7 |
9 |
Fold:TrkA C-terminal domain-like
Superfamily:TrkA C-terminal domain-like
Family:TrkA C-terminal domain-like |
| 33 | d1kbla2 |
|
not modelled |
12.1 |
11 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 34 | d1e8ca2 |
|
not modelled |
11.8 |
14 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 35 | c3a56B_ |
|
not modelled |
11.6 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein-glutaminase;
PDBTitle: crystal structure of pro- protein-glutaminase
|
| 36 | c3eatX_ |
|
not modelled |
11.5 |
13 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:pyoverdine biosynthesis protein pvcb;
PDBTitle: crystal structure of the pvcb (pa2255) protein from2 pseudomonas aeruginosa
|
| 37 | d2ptza1 |
|
not modelled |
11.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 38 | c2wbqA_ |
|
not modelled |
10.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arginine beta-hydroxylase;
PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
|
| 39 | d2gdwa1 |
|
not modelled |
10.6 |
21 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Peptidyl carrier domain |
| 40 | c3da5A_ |
|
not modelled |
10.5 |
14 |
PDB header:rna binding protein
Chain: A: PDB Molecule:argonaute;
PDBTitle: crystal structure of piwi/argonaute/zwille(paz) domain from2 thermococcus thioreducens
|
| 41 | d1wpna_ |
|
not modelled |
10.3 |
10 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 42 | d1vk3a3 |
|
not modelled |
10.2 |
15 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 43 | c1wynA_ |
|
not modelled |
9.8 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:calponin-2;
PDBTitle: solution structure of the ch domain of human calponin-2
|
| 44 | d1k20a_ |
|
not modelled |
9.6 |
15 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 45 | d2jfga2 |
|
not modelled |
9.5 |
19 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 46 | d1h6za2 |
|
not modelled |
9.4 |
18 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 47 | d1ds1a_ |
|
not modelled |
9.0 |
22 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Clavaminate synthase |
| 48 | c3pvjB_ |
|
not modelled |
8.9 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of the fe(ii)/alpha-ketoglutarate dependent taurine2 dioxygenase from pseudomonas putida kt2440
|
| 49 | c2og5A_ |
|
not modelled |
8.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxygenase;
PDBTitle: crystal structure of asparagine oxygenase (asno)
|
| 50 | d1twia1 |
|
not modelled |
8.9 |
9 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 51 | c2cmpA_ |
|
not modelled |
8.3 |
19 |
PDB header:terminase
Chain: A: PDB Molecule:terminase small subunit;
PDBTitle: crystal structure of the dna binding domain of g1p small2 terminase subunit from bacteriophage sf6
|
| 52 | d1otja_ |
|
not modelled |
8.3 |
27 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 53 | c3r1jB_ |
|
not modelled |
8.1 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
|
| 54 | c2lc3A_ |
|
not modelled |
7.9 |
30 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase hectd1;
PDBTitle: solution nmr structure of a helical bundle domain from human e3 ligase2 hectd1. northeast structural genomics consortium (nesg) target3 ht6305a
|
| 55 | d1oiha_ |
|
not modelled |
7.7 |
20 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 56 | c2wawA_ |
|
not modelled |
7.6 |
7 |
PDB header:unknown function
Chain: A: PDB Molecule:moba relate protein;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0371c2 homolog from mycobacterium sp. strain jc1
|
| 57 | c3nnlB_ |
|
not modelled |
7.2 |
20 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cura;
PDBTitle: halogenase domain from cura module (crystal form iii)
|
| 58 | c3n29A_ |
|
not modelled |
7.1 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:carboxynorspermidine decarboxylase;
PDBTitle: crystal structure of carboxynorspermidine decarboxylase complexed with2 norspermidine from campylobacter jejuni
|
| 59 | c2pfrB_ |
|
not modelled |
7.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:arylamine n-acetyltransferase 2;
PDBTitle: human n-acetyltransferase 2
|
| 60 | d1xjsa_ |
|
not modelled |
6.8 |
22 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 61 | d1jr7a_ |
|
not modelled |
6.5 |
7 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Gab protein (hypothetical protein YgaT) |
| 62 | d2hawa1 |
|
not modelled |
6.4 |
10 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 63 | c3bdeA_ |
|
not modelled |
6.4 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:mll5499 protein;
PDBTitle: crystal structure of a dabb family protein with a ferredoxin-like fold2 (mll5499) from mesorhizobium loti maff303099 at 1.79 a resolution
|
| 64 | d1hkva1 |
|
not modelled |
6.4 |
7 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 65 | c3bguA_ |
|
not modelled |
6.4 |
7 |
PDB header:unknown function
Chain: A: PDB Molecule:ferredoxin-like protein of unknown function;
PDBTitle: crystal structure of a dimeric ferredoxin-like protein of unknown2 function (tfu_0763) from thermobifida fusca yx at 1.50 a resolution
|
| 66 | c3al6A_ |
|
not modelled |
6.2 |
20 |
PDB header:unknown function
Chain: A: PDB Molecule:jmjc domain-containing protein c2orf60;
PDBTitle: crystal structure of human tyw5
|
| 67 | d2bsza1 |
|
not modelled |
6.1 |
21 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
| 68 | c3jxoB_ |
|
not modelled |
5.8 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:trka-n domain protein;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
| 69 | d1w6va1 |
|
not modelled |
5.8 |
25 |
Fold:DUSP-like
Superfamily:DUSP-like
Family:DUSP, domain in ubiquitin-specific proteases |
| 70 | c2cr4A_ |
|
not modelled |
5.7 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:sh3 domain-binding protein 2;
PDBTitle: solution structure of the sh2 domain of human sh3bp2 protein
|
| 71 | d1asua_ |
|
not modelled |
5.7 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 72 | c3lybC_ |
|
not modelled |
5.7 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative endoribonuclease;
PDBTitle: structure of putative endoribonuclease(kp1_3112) from2 klebsiella pneumoniae
|
| 73 | d1w4ta1 |
|
not modelled |
5.5 |
13 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
| 74 | c2r32A_ |
|
not modelled |
5.5 |
31 |
PDB header:immune system
Chain: A: PDB Molecule:gcn4-pii/tumor necrosis factor ligand
PDBTitle: crystal structure of human gitrl variant
|
| 75 | c3d01G_ |
|
not modelled |
5.5 |
23 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the protein atu1372 with unknown function from2 agrobacterium tumefaciens
|
| 76 | c2zkqq_ |
|
not modelled |
5.4 |
25 |
PDB header:ribosomal protein/rna
Chain: Q: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 77 | d1hyoa2 |
|
not modelled |
5.4 |
23 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 78 | c1nhgD_ |
|
not modelled |
5.3 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: crystal structure analysis of plasmodium falciparum enoyl-2 acyl-carrier-protein reductase with triclosan
|
| 79 | c2eobA_ |
|
not modelled |
5.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:1-phosphatidylinositol-4,5-bisphosphate
PDBTitle: solution structure of the second sh2 domain from rat plc2 gamma-2
|
| 80 | c1e8cB_ |
|
not modelled |
5.2 |
8 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide2 synthetase from e. coli
|
| 81 | c2r8uA_ |
|
not modelled |
5.2 |
9 |
PDB header:cell cycle
Chain: A: PDB Molecule:microtubule-associated protein rp/eb family
PDBTitle: structure of fragment of human end-binding protein 1 (eb1)2 containing the n-terminal domain at 1.35 a resolution
|
| 82 | d2hg6a1 |
|
not modelled |
5.2 |
67 |
Fold:PA1123-like
Superfamily:PA1123-like
Family:PA1123-like |
| 83 | d1r9pa_ |
|
not modelled |
5.1 |
19 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |