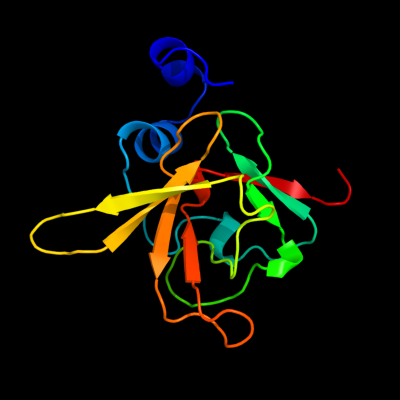
| 1 | d1l1da_
|
|
 |
100.0 |
47 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
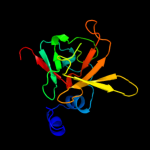
| 2 | c3e0mB_
|
|
 |
100.0 |
52 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb
PDBTitle: crystal structure of fusion protein of msra and msrb
|
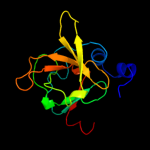
| 3 | c2k8dA_
|
|
 |
100.0 |
51 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msrb;
PDBTitle: solution structure of a zinc-binding methionine sulfoxide reductase
|
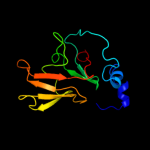
| 4 | c3hcjB_
|
|
 |
100.0 |
51 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: structure of msrb from xanthomonas campestris (oxidized2 form)
|
| 5 | d1xm0a1
|
|
 |
100.0 |
50 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
| 6 | c3cezA_
|
|
 |
100.0 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase;
PDBTitle: crystal structure of methionine-r-sulfoxide reductase from2 burkholderia pseudomallei
|
| 7 | c2l1uA_
|
|
 |
100.0 |
52 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b2, mitochondrial;
PDBTitle: structure-functional analysis of mammalian msrb2 protein
|
| 8 | c2kaoA_
|
|
 |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b1;
PDBTitle: structure of reduced mouse methionine sulfoxide reductase b12 (sec95cys mutant)
|
| 9 | c3facE_
|
|
 |
61.8 |
21 |
PDB header:unknown function
Chain: E: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of rhodobacter sphaeroides protein2 rsp_2168. northeast structural genomics target rhr83.
|
| 10 | d1aj7h1
|
|
 |
48.4 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 11 | d1gafh1
|
|
 |
46.1 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 12 | d1v7mh1
|
|
 |
45.6 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 13 | d1vera_
|
|
 |
41.5 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 14 | d1j8hd1
|
|
 |
41.4 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 15 | d1pfta_
|
|
 |
40.6 |
21 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 16 | d1b88a_
|
|
 |
36.8 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 17 | d1dl6a_
|
|
 |
35.9 |
24 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 18 | d1miml1
|
|
 |
35.5 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 19 | d1ggbh1
|
|
 |
33.8 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 20 | d1nakh1
|
|
 |
32.5 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 21 | c3goxB_ |
|
not modelled |
30.2 |
26 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:restriction endonuclease hpy99i;
PDBTitle: crystal structure of the beta-beta-alpha-me type ii restriction2 endonuclease hpy99i in the absence of edta
|
| 22 | d3dgga1 |
|
not modelled |
29.2 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 23 | c2w4rB_ |
|
not modelled |
28.6 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable atp-dependent rna helicase dhx58;
PDBTitle: crystal structure of the regulatory domain of human lgp2
|
| 24 | d1beca1 |
|
not modelled |
28.5 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 25 | d1dfbl1 |
|
not modelled |
28.5 |
24 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 26 | c1ua6H_ |
|
not modelled |
28.2 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant sfsf complexed with2 hen egg white lysozyme complex
|
| 27 | d1jnlh1 |
|
not modelled |
27.7 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 28 | d1w72h1 |
|
not modelled |
27.4 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 29 | c1ic4H_ |
|
not modelled |
26.6 |
20 |
PDB header:protein binding/hydrolase
Chain: H: PDB Molecule:igg1 fab chain h;
PDBTitle: crystal structure of hyhel-10 fv mutant(hd32a)-hen lysozyme2 complex
|
| 30 | c1s1iY_ |
|
not modelled |
26.2 |
42 |
PDB header:ribosome
Chain: Y: PDB Molecule:60s ribosomal protein l37-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 31 | c3h0gI_ |
|
not modelled |
26.2 |
18 |
PDB header:transcription
Chain: I: PDB Molecule:dna-directed rna polymerase ii subunit rpb9;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 32 | c2zkr2_ |
|
not modelled |
26.1 |
30 |
PDB header:ribosomal protein/rna
Chain: 2: PDB Molecule:60s ribosomal protein l37e;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 33 | d1smyd_ |
|
not modelled |
26.0 |
36 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
| 34 | c3k7aM_ |
|
not modelled |
25.7 |
20 |
PDB header:transcription
Chain: M: PDB Molecule:transcription initiation factor iib;
PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
|
| 35 | c1qokA_ |
|
not modelled |
25.1 |
22 |
PDB header:immunoglobulin
Chain: A: PDB Molecule:mfe-23 recombinant antibody fragment;
PDBTitle: mfe-23 an anti-carcinoembryonic antigen single-chain fv2 antibody
|
| 36 | d1lp9e1 |
|
not modelled |
25.0 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 37 | d1w72l1 |
|
not modelled |
25.0 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 38 | d1sm3h1 |
|
not modelled |
25.0 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 39 | c2dqeH_ |
|
not modelled |
24.7 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant (hy53a) complexed2 with hen egg lysozyme
|
| 40 | d1n7ml1 |
|
not modelled |
24.6 |
24 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 41 | d1dl7h_ |
|
not modelled |
24.5 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 42 | c2dqcH_ |
|
not modelled |
24.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant(hy33f) complexed2 with hen egg lysozyme
|
| 43 | d2oslb1 |
|
not modelled |
24.3 |
30 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 44 | c2znwB_ |
|
not modelled |
24.3 |
19 |
PDB header:immune system/hydrolase
Chain: B: PDB Molecule:scfv10;
PDBTitle: crystal structure of scfv10 in complex with hen egg lysozyme
|
| 45 | c1i3qI_ |
|
not modelled |
24.3 |
12 |
PDB header:transcription
Chain: I: PDB Molecule:dna-directed rna polymerase ii 14.2kd
PDBTitle: rna polymerase ii crystal form i at 3.1 a resolution
|
| 46 | d1ospl1 |
|
not modelled |
23.7 |
26 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 47 | d2ntsp1 |
|
not modelled |
23.7 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 48 | d1q9rb1 |
|
not modelled |
23.7 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 49 | d2i27n1 |
|
not modelled |
23.5 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 50 | d1nbyb1 |
|
not modelled |
23.5 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 51 | c4a18A_ |
|
not modelled |
23.2 |
29 |
PDB header:ribosome
Chain: A: PDB Molecule:ribosomal protein l37;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
|
| 52 | d1nsnl1 |
|
not modelled |
23.2 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 53 | d1pz5b1 |
|
not modelled |
23.2 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 54 | d1x6ma_ |
|
not modelled |
22.9 |
24 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:Glutathione-dependent formaldehyde-activating enzyme, Gfa |
| 55 | d2h1ph1 |
|
not modelled |
22.8 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 56 | d3c6la1 |
|
not modelled |
22.8 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 57 | d1igch1 |
|
not modelled |
22.7 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 58 | c3jywY_ |
|
not modelled |
22.4 |
42 |
PDB header:ribosome
Chain: Y: PDB Molecule:60s ribosomal protein l37(a);
PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
|
| 59 | c2gkiA_ |
|
not modelled |
22.4 |
30 |
PDB header:immune system
Chain: A: PDB Molecule:nuclease;
PDBTitle: heavy and light chain variable single domains of an anti-dna binding2 antibody hydrolyze both double- and single-stranded dnas without3 sequence specificity
|
| 60 | d1a4ka1 |
|
not modelled |
22.1 |
40 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 61 | d1nj9b1 |
|
not modelled |
21.9 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 62 | c3a6bH_ |
|
not modelled |
21.8 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh, anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant ln32d complexed with hen egg2 white lysozyme
|
| 63 | c2otuC_ |
|
not modelled |
21.8 |
16 |
PDB header:immune system
Chain: C: PDB Molecule:fv light chain variable domain;
PDBTitle: crystal structure of fv polyglutamine complex
|
| 64 | d1kcvh1 |
|
not modelled |
21.7 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 65 | d1ndmb1 |
|
not modelled |
21.7 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 66 | d1emth1 |
|
not modelled |
21.7 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 67 | d1igfl1 |
|
not modelled |
21.7 |
33 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 68 | c1dzbB_ |
|
not modelled |
21.6 |
26 |
PDB header:immune system
Chain: B: PDB Molecule:scfv fragment 1f9;
PDBTitle: crystal structure of phage library-derived single-chain fv2 fragment 1f9 in complex with turkey egg-white lysozyme
|
| 69 | c3cm9J_ |
|
not modelled |
21.6 |
30 |
PDB header:immune system
Chain: J: PDB Molecule:secretory component;
PDBTitle: solution structure of human siga2
|
| 70 | c3chnJ_ |
|
not modelled |
21.6 |
30 |
PDB header:immune system
Chain: J: PDB Molecule:secretory component;
PDBTitle: solution structure of human secretory iga1
|
| 71 | d1igil1 |
|
not modelled |
21.6 |
35 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 72 | c2dqdH_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant (hy50f) complexed2 with hen egg lysozyme
|
| 73 | c1c08B_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: B: PDB Molecule:anti-hen egg white lysozyme antibody (hyhel-10);
PDBTitle: crystal structure of hyhel-10 fv-hen lysozyme complex
|
| 74 | c2dqiH_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant (ly50a) complexed2 with hen egg lysozyme
|
| 75 | c3a67H_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh, anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant ln31d complexed with hen egg2 white lysozyme
|
| 76 | d1j1ph_ |
|
not modelled |
21.5 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 77 | c1j1pH_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant ls91a complexed2 with hen egg white lysozyme
|
| 78 | c3a6cH_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh, anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant ln92d complexed with hen egg2 white lysozyme
|
| 79 | c2dqjH_ |
|
not modelled |
21.5 |
20 |
PDB header:immune system/hydrolase
Chain: H: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv (wild-type) complexed with2 hen egg lysozyme at 1.8a resolution
|
| 80 | d2dd8l1 |
|
not modelled |
21.5 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 81 | d1gpoh1 |
|
not modelled |
21.2 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 82 | c3nn8B_ |
|
not modelled |
21.2 |
29 |
PDB header:immune system
Chain: B: PDB Molecule:engineered scfv, light chain;
PDBTitle: crystal structure of engineered antibody fragment based on 3d5
|
| 83 | d1dqlh_ |
|
not modelled |
21.1 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 84 | d1mamh1 |
|
not modelled |
21.0 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 85 | d1fpth1 |
|
not modelled |
20.9 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 86 | c1ic5H_ |
|
not modelled |
20.7 |
15 |
PDB header:protein binding/hydrolase
Chain: H: PDB Molecule:igg1 fab chain h;
PDBTitle: crystal structure of hyhel-10 fv mutant(hd99a)-hen lysozyme2 complex
|
| 87 | c2dqfE_ |
|
not modelled |
20.6 |
20 |
PDB header:immune system/hydrolase
Chain: E: PDB Molecule:ig vh,anti-lysozyme;
PDBTitle: crystal structure of hyhel-10 fv mutant (y33ay53a)2 complexed with hen egg lysozyme
|
| 88 | d1ub5b1 |
|
not modelled |
20.5 |
40 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 89 | d1a5fh1 |
|
not modelled |
20.3 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 90 | c1nqbC_ |
|
not modelled |
20.2 |
32 |
PDB header:immunoglobulin
Chain: C: PDB Molecule:single-chain antibody fragment;
PDBTitle: trivalent antibody fragment
|
| 91 | d1q9wb1 |
|
not modelled |
20.1 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 92 | d1h8ba_ |
|
not modelled |
20.1 |
30 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:EF-hand modules in multidomain proteins |
| 93 | d2qqnh1 |
|
not modelled |
20.0 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 94 | d1mexh1 |
|
not modelled |
19.9 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 95 | d1bbjb1 |
|
not modelled |
19.9 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 96 | c3trzE_ |
|
not modelled |
19.6 |
29 |
PDB header:rna binding protein/rna
Chain: E: PDB Molecule:protein lin-28 homolog a;
PDBTitle: mouse lin28a in complex with let-7d microrna pre-element
|
| 97 | d1kelh1 |
|
not modelled |
19.6 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 98 | d1l7tl1 |
|
not modelled |
19.5 |
38 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 99 | d2dkta2 |
|
not modelled |
19.5 |
31 |
Fold:Zinc hairpin stack
Superfamily:Zinc hairpin stack
Family:Zinc hairpin stack |