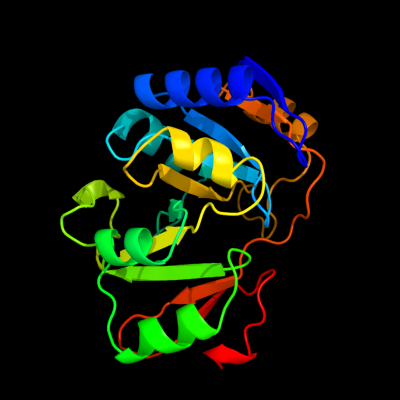
| 1 | d1k7ja_
|
|
 |
100.0 |
97 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:YrdC-like |
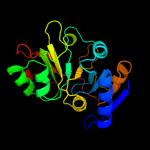
| 2 | c2eqaA_
|
|
 |
100.0 |
28 |
PDB header:rna binding protein
Chain: A: PDB Molecule:hypothetical protein st1526;
PDBTitle: crystal structure of the hypothetical sua5 protein from2 sulfolobus tokodaii
|
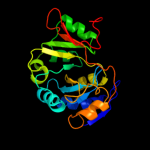
| 3 | d1jcua_
|
|
 |
100.0 |
30 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:YrdC-like |
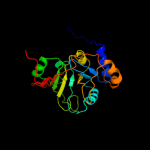
| 4 | d1hrua_
|
|
 |
100.0 |
23 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:YrdC-like |
| 5 | c3l7vA_
|
|
 |
100.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:putative uncharacterized protein smu.1377c;
PDBTitle: crystal structure of a hypothetical protein smu.1377c from2 streptococcus mutans ua159
|
| 6 | c3tsuA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: crystal structure of e. coli hypf with amp-pnp and carbamoyl phosphate
|
| 7 | d1t5la1
|
|
 |
81.0 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 8 | c2frxD_
|
|
 |
52.7 |
28 |
PDB header:transferase
Chain: D: PDB Molecule:hypothetical protein yebu;
PDBTitle: crystal structure of yebu, a m5c rna methyltransferase from e.coli
|
| 9 | d1d4oa_
|
|
 |
48.9 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
| 10 | c1pt9B_
|
|
 |
48.5 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase, mitochondrial;
PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
|
| 11 | c3m4xA_
|
|
 |
48.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:nol1/nop2/sun family protein;
PDBTitle: structure of a ribosomal methyltransferase
|
| 12 | c3m6wA_
|
|
 |
43.3 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:rrna methylase;
PDBTitle: multi-site-specific 16s rrna methyltransferase rsmf from thermus2 thermophilus in space group p21212 in complex with s-adenosyl-l-3 methionine
|
| 13 | c2bruC_
|
|
 |
42.8 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad(p) transhydrogenase subunit beta;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
| 14 | d1pnoa_
|
|
 |
41.7 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
| 15 | c2oemA_
|
|
 |
41.5 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate enolase;
PDBTitle: crystal structure of a rubisco-like protein from geobacillus2 kaustophilus liganded with mg2+ and 2,3-diketohexane 1-phosphate
|
| 16 | d1hq0a_
|
|
 |
40.9 |
26 |
Fold:CNF1/YfiH-like putative cysteine hydrolases
Superfamily:CNF1/YfiH-like putative cysteine hydrolases
Family:Type 1 cytotoxic necrotizing factor, catalytic domain |
| 17 | d1ixka_
|
|
 |
37.1 |
28 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:NOL1/NOP2/sun |
| 18 | c3fk4A_
|
|
 |
36.3 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:rubisco-like protein;
PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
|
| 19 | c3p3dA_
|
|
 |
35.2 |
32 |
PDB header:nuclear protein
Chain: A: PDB Molecule:nucleoporin 53;
PDBTitle: crystal structure of the nup53 rrm domain from pichia guilliermondii
|
| 20 | c3nwrA_
|
|
 |
33.8 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
| 21 | d1ykwa1 |
|
not modelled |
32.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 22 | d2d69a1 |
|
not modelled |
32.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 23 | c2zviB_ |
|
not modelled |
32.3 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate
PDBTitle: crystal structure of 2,3-diketo-5-methylthiopentyl-1-2 phosphate enolase from bacillus subtilis
|
| 24 | d1sqga2 |
|
not modelled |
32.0 |
23 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:NOL1/NOP2/sun |
| 25 | d1rbla1 |
|
not modelled |
31.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 26 | d1bxna1 |
|
not modelled |
30.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 27 | d1ej7l1 |
|
not modelled |
29.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 28 | c2qj8B_ |
|
not modelled |
29.1 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:mlr6093 protein;
PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
|
| 29 | c2qs0A_ |
|
not modelled |
27.5 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:quinolinate synthetase a;
PDBTitle: quinolinate synthase from pyrococcus furiosus
|
| 30 | c3qfwB_ |
|
not modelled |
27.1 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase large
PDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
|
| 31 | c3mioA_ |
|
not modelled |
27.0 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:3,4-dihydroxy-2-butanone 4-phosphate synthase;
PDBTitle: crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase2 domain from mycobacterium tuberculosis at ph 6.00
|
| 32 | d1gk8a1 |
|
not modelled |
26.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 33 | c2yxlA_ |
|
not modelled |
25.5 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:450aa long hypothetical fmu protein;
PDBTitle: crystal structure of ph0851
|
| 34 | c1sqgA_ |
|
not modelled |
25.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:sun protein;
PDBTitle: the crystal structure of the e. coli fmu apoenzyme at 1.652 a resolution
|
| 35 | d1svda1 |
|
not modelled |
25.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 36 | c3hjhA_ |
|
not modelled |
25.3 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair-coupling factor;
PDBTitle: a rigid n-terminal clamp restrains the motor domains of the bacterial2 transcription-repair coupling factor
|
| 37 | c3a4tA_ |
|
not modelled |
23.7 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase mj0026;
PDBTitle: crystal structure of atrm4 from m.jannaschii with sinefungin
|
| 38 | c3axtA_ |
|
not modelled |
23.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:probable n(2),n(2)-dimethylguanosine trna methyltransferase
PDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
|
| 39 | c1solA_ |
|
not modelled |
22.9 |
7 |
PDB header:actin-binding protein
Chain: A: PDB Molecule:gelsolin (150-169);
PDBTitle: a pip2 and f-actin-binding site of gelsolin, residue 150-2 169 (nmr, averaged structure)
|
| 40 | d1c4oa1 |
|
not modelled |
22.8 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 41 | d1geha1 |
|
not modelled |
22.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 42 | c1rldB_ |
|
not modelled |
21.2 |
15 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose 1,5 bisphosphate carboxylase/oxygenase (large
PDBTitle: solid-state phase transition in the crystal structure of ribulose 1,5-2 biphosphate carboxylase(slash)oxygenase
|
| 43 | d1snna_ |
|
not modelled |
21.2 |
15 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 44 | d2dula1 |
|
not modelled |
21.0 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TRM1-like |
| 45 | c2d69B_ |
|
not modelled |
20.5 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
| 46 | d1r0ua_ |
|
not modelled |
20.1 |
20 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Hypothetical protein YwiB |
| 47 | c2qygC_ |
|
not modelled |
18.7 |
13 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
| 48 | c1rcxH_ |
|
not modelled |
17.6 |
15 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
| 49 | c1telA_ |
|
not modelled |
16.9 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
| 50 | d1bwva1 |
|
not modelled |
16.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 51 | c2gq1A_ |
|
not modelled |
14.7 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:fructose-1,6-bisphosphatase;
PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
|
| 52 | d8ruca1 |
|
not modelled |
14.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 53 | c1gehE_ |
|
not modelled |
14.5 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase;
PDBTitle: crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate2 carboxylase/oxygenase)
|
| 54 | c2fhyL_ |
|
not modelled |
14.0 |
12 |
PDB header:hydrolase
Chain: L: PDB Molecule:fructose-1,6-bisphosphatase 1;
PDBTitle: structure of human liver fpbase complexed with a novel2 benzoxazole as allosteric inhibitor
|
| 55 | d2eyqa4 |
|
not modelled |
14.0 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 56 | c3n0vD_ |
|
not modelled |
12.9 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 57 | c3p04A_ |
|
not modelled |
12.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized bcr;
PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
|
| 58 | d1tksa_ |
|
not modelled |
12.0 |
21 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 59 | c3ckkA_ |
|
not modelled |
11.6 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanine-n(7)-)-methyltransferase;
PDBTitle: crystal structure of human methyltransferase-like protein 1
|
| 60 | d1nuwa_ |
|
not modelled |
11.6 |
16 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 61 | d1k4ia_ |
|
not modelled |
11.6 |
15 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 62 | c3louB_ |
|
not modelled |
11.5 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 63 | c1kevB_ |
|
not modelled |
11.4 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: structure of nadp-dependent alcohol dehydrogenase
|
| 64 | d1wdda1 |
|
not modelled |
10.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 65 | c2ekgB_ |
|
not modelled |
10.6 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:proline dehydrogenase/delta-1-pyrroline-5-carboxylate
PDBTitle: structure of thermus thermophilus proline dehydrogenase inactivated by2 n-propargylglycine
|
| 66 | d1bhea_ |
|
not modelled |
10.4 |
15 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:Galacturonase |
| 67 | c2eyqA_ |
|
not modelled |
9.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
|
| 68 | c2vduE_ |
|
not modelled |
9.9 |
25 |
PDB header:transferase
Chain: E: PDB Molecule:trna (guanine-n(7)-)-methyltransferase;
PDBTitle: structure of trm8-trm82, the yeast trna m7g methylation2 complex
|
| 69 | d1ftaa_ |
|
not modelled |
9.8 |
12 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 70 | c3crw1_ |
|
not modelled |
9.3 |
29 |
PDB header:hydrolase
Chain: 1: PDB Molecule:xpd/rad3 related dna helicase;
PDBTitle: "xpd_apo"
|
| 71 | c1bwvA_ |
|
not modelled |
8.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ribulose bisphosphate carboxylase);
PDBTitle: activated ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco)2 complexed with the reaction intermediate analogue 2-carboxyarabinitol3 1,5-bisphosphate
|
| 72 | d1d9qa_ |
|
not modelled |
8.6 |
22 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 73 | d1duvg1 |
|
not modelled |
8.3 |
17 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 74 | d1h80a_ |
|
not modelled |
8.2 |
18 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:iota-carrageenase |
| 75 | d1q32a2 |
|
not modelled |
8.2 |
44 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 76 | d2hi6a1 |
|
not modelled |
8.1 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:AF0055-like |
| 77 | d1g57a_ |
|
not modelled |
7.7 |
12 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 78 | d2f06a1 |
|
not modelled |
7.7 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 79 | c3p04B_ |
|
not modelled |
7.6 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized bcr;
PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
|
| 80 | d1knza_ |
|
not modelled |
7.5 |
17 |
Fold:NSP3 homodimer
Superfamily:NSP3 homodimer
Family:NSP3 homodimer |
| 81 | c2ouiB_ |
|
not modelled |
7.2 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: d275p mutant of alcohol dehydrogenase from protozoa entamoeba2 histolytica
|
| 82 | d1spia_ |
|
not modelled |
7.2 |
24 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 83 | d1bk4a_ |
|
not modelled |
7.1 |
16 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 84 | d1yksa2 |
|
not modelled |
7.1 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RNA helicase |
| 85 | c2y75F_ |
|
not modelled |
7.1 |
25 |
PDB header:transcription
Chain: F: PDB Molecule:hth-type transcriptional regulator cymr;
PDBTitle: the structure of cymr (yrzc) the global cysteine regulator2 of b. subtilis
|
| 86 | d1xcfa_ |
|
not modelled |
7.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 87 | c3d7aB_ |
|
not modelled |
7.0 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0201 protein ph1010;
PDBTitle: crystal structure of duf54 family protein ph1010 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
|
| 88 | c1f8fA_ |
|
not modelled |
7.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:benzyl alcohol dehydrogenase;
PDBTitle: crystal structure of benzyl alcohol dehydrogenase from acinetobacter2 calcoaceticus
|
| 89 | d1otha1 |
|
not modelled |
6.9 |
16 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 90 | d1tg7a3 |
|
not modelled |
6.7 |
18 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Beta-galactosidase LacA, domains 4 and 5 |
| 91 | d1jy1a2 |
|
not modelled |
6.6 |
44 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 92 | c1r37B_ |
|
not modelled |
6.4 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase from sulfolobus solfataricus2 complexed with nad(h) and 2-ethoxyethanol
|
| 93 | d1zq1a2 |
|
not modelled |
6.3 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 94 | d1yzha1 |
|
not modelled |
6.2 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TrmB-like |
| 95 | d1mvfd_ |
|
not modelled |
6.0 |
38 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:Kis/PemI addiction antidote |
| 96 | d2b2na1 |
|
not modelled |
5.9 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 97 | d1vlva1 |
|
not modelled |
5.8 |
23 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 98 | c2v34B_ |
|
not modelled |
5.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:4-diphosphocytidyl-2c-methyl-d-erythritol kinase;
PDBTitle: ispe in complex with cytidine and ligand
|
| 99 | c3h1tA_ |
|
not modelled |
5.7 |
44 |
PDB header:hydrolase
Chain: A: PDB Molecule:type i site-specific restriction-modification
PDBTitle: the fragment structure of a putative hsdr subunit of a type2 i restriction enzyme from vibrio vulnificus yj016
|