| 1 | c3nzkB_ |
|
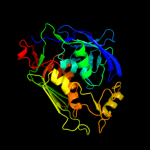 |
100.0 |
93 |
PDB header:hydrolase
Chain: B: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] n-acetylglucosamine
PDBTitle: structure of lpxc from yersinia enterocolitica complexed with chir0902 inhibitor
|
| 2 | c2vesA_ |
|
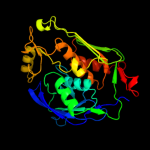 |
100.0 |
59 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] n-acetylglucosamine
PDBTitle: crystal structure of lpxc from pseudomonas aeruginosa2 complexed with the potent bb-78485 inhibitor
|
| 3 | c2go4A_ |
|
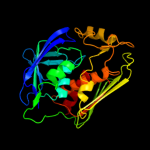 |
100.0 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] n-acetylglucosamine
PDBTitle: crystal structure of aquifex aeolicus lpxc complexed with tu-514
|
| 4 | d1p42a2 |
|
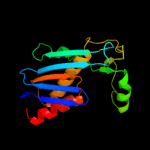 |
100.0 |
33 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC |
| 5 | d1p42a1 |
|
 |
100.0 |
33 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC |
| 6 | d2bv3a3 |
|
 |
86.3 |
17 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 7 | d1pkpa1 |
|
 |
66.7 |
47 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 8 | d2uube1 |
|
 |
65.3 |
47 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 9 | c2zkqe_ |
|
 |
55.1 |
47 |
PDB header:ribosomal protein/rna
Chain: E: PDB Molecule:rna expansion segment es6 part ii;
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 10 | d2qale1 |
|
 |
53.5 |
33 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 11 | c1s1hE_ |
|
 |
52.7 |
47 |
PDB header:ribosome
Chain: E: PDB Molecule:40s ribosomal protein s2;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 12 | c3bbnE_ |
|
 |
50.7 |
53 |
PDB header:ribosome
Chain: E: PDB Molecule:ribosomal protein s5;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 13 | d1u5tb2 |
|
 |
45.6 |
50 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 14 | d2azea1 |
|
 |
45.4 |
25 |
Fold:E2F-DP heterodimerization region
Superfamily:E2F-DP heterodimerization region
Family:DP dimerization segment |
| 15 | c2zkru_ |
|
 |
43.2 |
31 |
PDB header:ribosomal protein/rna
Chain: U: PDB Molecule:rna expansion segment es41;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 16 | d1vqou1 |
|
 |
41.1 |
19 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:Ribosomal protein L24e |
| 17 | c2xzmE_ |
|
 |
41.1 |
47 |
PDB header:ribosome
Chain: E: PDB Molecule:ribosomal protein s5 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 18 | c3izcZ_ |
|
 |
41.0 |
19 |
PDB header:ribosome
Chain: Z: PDB Molecule:60s ribosomal protein rpl24 (l24e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 19 | c4a1eT_ |
|
 |
40.8 |
13 |
PDB header:ribosome
Chain: T: PDB Molecule:rpl24;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
|
| 20 | c3ccjU_ |
|
 |
40.6 |
19 |
PDB header:ribosome
Chain: U: PDB Molecule:50s ribosomal protein l24e;
PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation c2534u
|
| 21 | c1eg0B_ |
|
not modelled |
39.4 |
47 |
PDB header:ribosome
PDB COMPND:
|
| 22 | c3iz5Z_ |
|
not modelled |
39.3 |
19 |
PDB header:ribosome
Chain: Z: PDB Molecule:60s ribosomal protein l24 (l24e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 23 | c1p6gE_ |
|
not modelled |
36.9 |
33 |
PDB header:ribosome
Chain: E: PDB Molecule:30s ribosomal protein s5;
PDBTitle: real space refined coordinates of the 30s subunit fitted2 into the low resolution cryo-em map of the ef-g.gtp state3 of e. coli 70s ribosome
|
| 24 | d1j6xa_ |
|
not modelled |
29.7 |
25 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 25 | d2hi6a1 |
|
not modelled |
28.4 |
20 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:AF0055-like |
| 26 | d1w07a3 |
|
not modelled |
27.8 |
12 |
Fold:Acyl-CoA dehydrogenase NM domain-like
Superfamily:Acyl-CoA dehydrogenase NM domain-like
Family:acyl-CoA oxidase N-terminal domains |
| 27 | d1j98a_ |
|
not modelled |
27.1 |
55 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 28 | c1s1iS_ |
|
not modelled |
25.9 |
19 |
PDB header:ribosome
Chain: S: PDB Molecule:60s ribosomal protein l24-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 29 | d1l4db_ |
|
not modelled |
25.7 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Staphylokinase/streptokinase
Family:Staphylokinase/streptokinase |
| 30 | d1vjea_ |
|
not modelled |
24.8 |
35 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 31 | d1j6wa_ |
|
not modelled |
22.4 |
29 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 32 | d2dy1a3 |
|
not modelled |
22.2 |
19 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 33 | c2ddhA_ |
|
not modelled |
19.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:acyl-coa oxidase;
PDBTitle: crystal structure of acyl-coa oxidase complexed with 3-oh-dodecanoate
|
| 34 | d1bmlc1 |
|
not modelled |
18.2 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Staphylokinase/streptokinase
Family:Staphylokinase/streptokinase |
| 35 | c1w07A_ |
|
not modelled |
17.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:acyl-coa oxidase;
PDBTitle: arabidopsis thaliana acyl-coa oxidase 1
|
| 36 | c2ow8f_ |
|
not modelled |
16.6 |
47 |
PDB header:ribosome
Chain: F: PDB Molecule:
PDBTitle: crystal structure of a 70s ribosome-trna complex reveals functional2 interactions and rearrangements. this file, 2ow8, contains the 30s3 ribosome subunit, two trna, and mrna molecules. 50s ribosome subunit4 is in the file 1vsa.
|
| 37 | d1gsoa2 |
|
not modelled |
15.2 |
22 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 38 | d1l4zb_ |
|
not modelled |
14.5 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Staphylokinase/streptokinase
Family:Staphylokinase/streptokinase |
| 39 | d2pw9a1 |
|
not modelled |
14.2 |
15 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:FdhD/NarQ |
| 40 | d1vh1a_ |
|
not modelled |
14.1 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 41 | c2fonA_ |
|
not modelled |
13.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peroxisomal acyl-coa oxidase 1a;
PDBTitle: x-ray crystal structure of leacx1, an acyl-coa oxidase from2 lycopersicon esculentum (tomato)
|
| 42 | d2ddha3 |
|
not modelled |
13.4 |
9 |
Fold:Acyl-CoA dehydrogenase NM domain-like
Superfamily:Acyl-CoA dehydrogenase NM domain-like
Family:acyl-CoA oxidase N-terminal domains |
| 43 | c3izbE_ |
|
not modelled |
13.2 |
47 |
PDB header:ribosome
Chain: E: PDB Molecule:40s ribosomal protein rps2 (s5p);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 44 | c2kyzA_ |
|
not modelled |
13.0 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:heavy metal binding protein;
PDBTitle: nmr structure of heavy metal binding protein tm0320 from thermotoga2 maritima
|
| 45 | c3iz6E_ |
|
not modelled |
13.0 |
40 |
PDB header:ribosome
Chain: E: PDB Molecule:40s ribosomal protein s2 (s5p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 46 | c3m05A_ |
|
not modelled |
12.9 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pepe_1480;
PDBTitle: the crystal structure of a functionally unknown protein2 pepe_1480 from pediococcus pentosaceus atcc 25745
|
| 47 | d1pvxa_ |
|
not modelled |
12.5 |
23 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 48 | d1v54i_ |
|
not modelled |
12.4 |
26 |
Fold:Single transmembrane helix
Superfamily:Mitochondrial cytochrome c oxidase subunit VIc
Family:Mitochondrial cytochrome c oxidase subunit VIc |
| 49 | d1vh3a_ |
|
not modelled |
12.2 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 50 | d1vaea_ |
|
not modelled |
11.9 |
19 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 51 | c2yuyA_ |
|
not modelled |
11.2 |
22 |
PDB header:signaling protein
Chain: A: PDB Molecule:rho gtpase activating protein 21;
PDBTitle: solution structure of pdz domain of rho gtpase activating2 protein 21
|
| 52 | d1ky9b2 |
|
not modelled |
11.0 |
24 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 53 | c3m0dC_ |
|
not modelled |
10.6 |
30 |
PDB header:signaling protein
Chain: C: PDB Molecule:tnf receptor-associated factor 1;
PDBTitle: crystal structure of the traf1:traf2:ciap2 complex
|
| 54 | d1gpca_ |
|
not modelled |
10.5 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Phage ssDNA-binding proteins |
| 55 | c2vsvB_ |
|
not modelled |
10.1 |
15 |
PDB header:protein-binding
Chain: B: PDB Molecule:rhophilin-2;
PDBTitle: crystal structure of the pdz domain of human rhophilin-2
|
| 56 | c2lfcA_ |
|
not modelled |
10.1 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase, flavoprotein subunit;
PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
|
| 57 | d1u3ba2 |
|
not modelled |
10.1 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 58 | d1y0pa3 |
|
not modelled |
9.9 |
42 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 59 | d1s1ma2 |
|
not modelled |
9.3 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 60 | c1n0fF_ |
|
not modelled |
9.1 |
11 |
PDB header:biosynthetic protein
Chain: F: PDB Molecule:protein mraz;
PDBTitle: crystal structure of a cell division and cell wall2 biosynthesis protein upf0040 from mycoplasma pneumoniae:3 indication of a novel fold with a possible new conserved4 sequence motif
|
| 61 | d1v97a2 |
|
not modelled |
9.0 |
39 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 62 | c2krgA_ |
|
not modelled |
8.7 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf1;
PDBTitle: solution structure of human sodium/ hydrogen exchange2 regulatory factor 1(150-358)
|
| 63 | c2qdrA_ |
|
not modelled |
8.5 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative dioxygenase (npun_f5605) from nostoc2 punctiforme pcc 73102 at 2.60 a resolution
|
| 64 | c2dcoA_ |
|
not modelled |
8.4 |
56 |
PDB header:membrane protein
Chain: A: PDB Molecule:s1p4 first extracellular loop peptidomimetic;
PDBTitle: s1p4 first extracellular loop peptidomimetic
|
| 65 | d1nz8a_ |
|
not modelled |
8.3 |
22 |
Fold:Ferredoxin-like
Superfamily:N-utilization substance G protein NusG, N-terminal domain
Family:N-utilization substance G protein NusG, N-terminal domain |
| 66 | c2kr7A_ |
|
not modelled |
8.2 |
32 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase slyd;
PDBTitle: solution structure of helicobacter pylori slyd
|
| 67 | d1xpma2 |
|
not modelled |
8.1 |
18 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Chalcone synthase-like |
| 68 | d1qo8a3 |
|
not modelled |
8.0 |
32 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 69 | d1n62a2 |
|
not modelled |
8.0 |
27 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 70 | d1nfpa_ |
|
not modelled |
8.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
| 71 | c3smaD_ |
|
not modelled |
7.9 |
26 |
PDB header:transferase
Chain: D: PDB Molecule:frbf;
PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
|
| 72 | c2ad5B_ |
|
not modelled |
7.9 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: mechanisms of feedback regulation and drug resistance of ctp2 synthetases: structure of the e. coli ctps/ctp complex at 2.8-3 angstrom resolution.
|
| 73 | d1ynaa_ |
|
not modelled |
7.8 |
16 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 74 | d1gpma2 |
|
not modelled |
7.6 |
23 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 75 | d1jvna1 |
|
not modelled |
7.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 76 | d1d4ca3 |
|
not modelled |
7.5 |
32 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
| 77 | c2qktB_ |
|
not modelled |
7.4 |
15 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:inactivation-no-after-potential d protein;
PDBTitle: crystal structure of the 5th pdz domain of inad
|
| 78 | c4a8aI_ |
|
not modelled |
7.2 |
35 |
PDB header:hydrolase/hydrolase
Chain: I: PDB Molecule:periplasmic ph-dependent serine endoprotease degq;
PDBTitle: asymmetric cryo-em reconstruction of e. coli degq 12-mer in complex2 with lysozyme
|
| 79 | d1vq2a_ |
|
not modelled |
7.2 |
22 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
| 80 | c2hfpB_ |
|
not modelled |
7.2 |
80 |
PDB header:transcription
Chain: B: PDB Molecule:src peptide fragment;
PDBTitle: crystal structure of ppar gamma with n-sulfonyl-2-indole2 carboxamide ligands
|
| 81 | c3kmgE_ |
|
not modelled |
7.1 |
80 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator-1;
PDBTitle: the x-ray crystal structure of ppar-gamma in complex with an indole2 derivative modulator, gsk538, and an src-1 peptide
|
| 82 | c2x7jA_ |
|
not modelled |
7.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: structure of the menaquinone biosynthesis protein mend from2 bacillus subtilis
|
| 83 | c1fm6E_ |
|
not modelled |
7.1 |
80 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and rosiglitazone and co-activator peptides.
|
| 84 | c1k74E_ |
|
not modelled |
7.1 |
80 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.3 angstrom resolution crystal structure of the2 heterodimer of the human ppargamma and rxralpha ligand3 binding domains respectively bound with gw409544 and 9-cis4 retinoic acid and co-activator peptides.
|
| 85 | c1fm9E_ |
|
not modelled |
7.1 |
80 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and gi262570 and co-activator peptides.
|
| 86 | d1rm6c2 |
|
not modelled |
7.0 |
36 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 87 | c1p8dC_ |
|
not modelled |
7.0 |
80 |
PDB header:membrane protein/protein binding
Chain: C: PDB Molecule:nuclear receptor coactivator 1 isoform 3;
PDBTitle: x-ray crystal structure of lxr ligand binding domain with 24(s),25-2 epoxycholesterol
|
| 88 | d1ukka_ |
|
not modelled |
7.0 |
40 |
Fold:OsmC-like
Superfamily:OsmC-like
Family:Ohr/OsmC resistance proteins |
| 89 | c1fm6V_ |
|
not modelled |
7.0 |
80 |
PDB header:transcription
Chain: V: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and rosiglitazone and co-activator peptides.
|
| 90 | d1vcoa2 |
|
not modelled |
7.0 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 91 | c3ipqB_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: B: PDB Molecule:nuclear receptor coactivator 1;
PDBTitle: x-ray structure of gw3965 synthetic agonist bound to the lxr-alpha
|
| 92 | c3ipuD_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: D: PDB Molecule:nuclear receptor coactivator 1;
PDBTitle: x-ray structure of benzisoxazole urea synthetic agonist bound to the2 lxr-alpha
|
| 93 | c1nrlC_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: C: PDB Molecule:nuclear receptor coactivator 1 isoform 3;
PDBTitle: crystal structure of the human pxr-lbd in complex with an2 src-1 coactivator peptide and sr12813
|
| 94 | c3ipsD_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: D: PDB Molecule:nuclear receptor coactivator 1;
PDBTitle: x-ray structure of benzisoxazole synthetic agonist bound to the lxr-2 alpha
|
| 95 | c3ipuC_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: C: PDB Molecule:nuclear receptor coactivator 1;
PDBTitle: x-ray structure of benzisoxazole urea synthetic agonist bound to the2 lxr-alpha
|
| 96 | c3ipsC_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: C: PDB Molecule:nuclear receptor coactivator 1;
PDBTitle: x-ray structure of benzisoxazole synthetic agonist bound to the lxr-2 alpha
|
| 97 | c1nrlD_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: D: PDB Molecule:nuclear receptor coactivator 1 isoform 3;
PDBTitle: crystal structure of the human pxr-lbd in complex with an2 src-1 coactivator peptide and sr12813
|
| 98 | c1fm6Y_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: Y: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and rosiglitazone and co-activator peptides.
|
| 99 | c1k7lB_ |
|
not modelled |
6.9 |
80 |
PDB header:transcription
Chain: B: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.5 angstrom resolution crystal structure of the human2 pparalpha ligand binding domain bound with gw409544 and a3 co-activator peptide.
|

