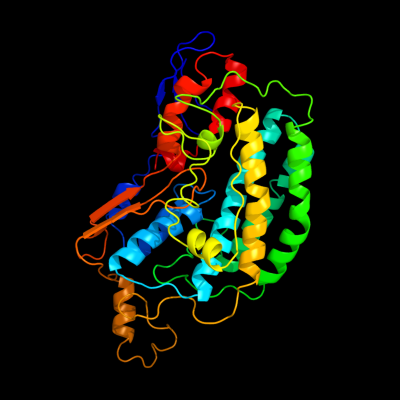
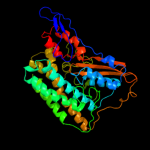
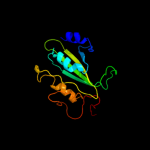
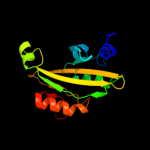
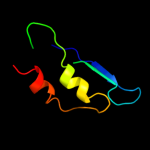
1 d2fug41
100.0
42
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nqo4-like2 c2fug4_
100.0
42
PDB header: oxidoreductaseChain: 4: PDB Molecule: nadh-quinone oxidoreductase chain 4;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
3 d1frfl_
100.0
15
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit4 d1e3db_
100.0
16
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit5 d1yq9h1
100.0
15
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit6 c1h2aL_
100.0
14
PDB header: oxidoreductaseChain: L: PDB Molecule: hydrogenase;PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
7 c2wpnB_
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: periplasmic [nifese] hydrogenase, large subunit,PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase2 from d. vulgaris hildenborough
8 d1cc1l_
100.0
17
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit9 c3myrB_
100.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: nickel-dependent hydrogenase large subunit;PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
10 d1wuil1
100.0
15
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit11 d2fug51
100.0
34
Fold: Nqo5-likeSuperfamily: Nqo5-likeFamily: Nqo5-like12 c3mcrA_
100.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh dehydrogenase, subunit c;PDBTitle: crystal structure of nadh dehydrogenase subunit c (tfu_2693) from2 thermobifida fusca yx-er1 at 2.65 a resolution
13 d1v97a4
45.2
2
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like14 c2ibpB_
29.2
15
PDB header: transferaseChain: B: PDB Molecule: citrate synthase;PDBTitle: crystal structure of citrate synthase from pyrobaculum aerophilum
15 d1ghha_
25.7
15
Fold: DNA damage-inducible protein DinISuperfamily: DNA damage-inducible protein DinIFamily: DNA damage-inducible protein DinI16 d1ffvc1
24.6
10
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like17 d2ntka1
23.4
23
Fold: Ntn hydrolase-likeSuperfamily: Archaeal IMP cyclohydrolase PurOFamily: Archaeal IMP cyclohydrolase PurO18 c1ufiD_
23.3
40
PDB header: dna binding proteinChain: D: PDB Molecule: major centromere autoantigen b;PDBTitle: crystal structure of the dimerization domain of human cenp-b
19 d1ufia_
23.0
40
Fold: ROP-likeSuperfamily: Dimerisation domain of CENP-BFamily: Dimerisation domain of CENP-B20 d2qswa1
21.4
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: NIL domain-like21 d2zpya3
not modelled
20.3
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM22 d1ni2a3
not modelled
19.6
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM23 d2qrra1
not modelled
19.4
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: NIL domain-like24 d1ef1a3
not modelled
19.0
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM25 c2w3zA_
not modelled
18.6
7
PDB header: hydrolaseChain: A: PDB Molecule: putative deacetylase;PDBTitle: structure of a streptococcus mutans ce4 esterase
26 d2c5sa2
not modelled
18.4
10
Fold: THUMP domainSuperfamily: THUMP domain-likeFamily: THUMP domain27 d1vqza1
not modelled
16.8
17
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SP1160 C-terminal domain-like28 c2dgyA_
not modelled
16.6
9
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
29 c3kfwX_
not modelled
16.0
36
PDB header: structural genomics, unknown functionChain: X: PDB Molecule: uncharacterized protein;PDBTitle: uncharacterized protein rv0674 from mycobacterium tuberculosis
30 c2f3oB_
not modelled
15.9
18
PDB header: unknown functionChain: B: PDB Molecule: pyruvate formate-lyase 2;PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
31 c2g40A_
not modelled
15.4
29
PDB header: unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a duf162 family protein (dr_1909) from2 deinococcus radiodurans at 1.70 a resolution
32 d2g40a1
not modelled
15.4
29
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: YkgG-like33 d1u07a_
not modelled
15.4
7
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB34 c3kwlA_
not modelled
15.3
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein from helicobacter pylori
35 d1gg3a3
not modelled
15.2
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM36 c2cveA_
not modelled
14.4
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1053;PDBTitle: crystal structure of a conserved hypothetical protein tt1547 from2 thermus thermophilus hb8
37 c3hrdC_
not modelled
14.1
12
PDB header: oxidoreductaseChain: C: PDB Molecule: nicotinate dehydrogenase fad-subunit;PDBTitle: crystal structure of nicotinate dehydrogenase
38 c2dnwA_
not modelled
12.9
17
PDB header: transport proteinChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of rsgi ruh-059, an acp domain of acyl2 carrier protein, mitochondrial [precursor] from human cdna
39 c2oqkA_
not modelled
12.8
6
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
40 c1wvtA_
not modelled
12.6
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein st2180;PDBTitle: crystal structure of uncharacterized protein st2180 from sulfolobus2 tokodaii
41 c2grxC_
not modelled
12.0
4
PDB header: metal transportChain: C: PDB Molecule: protein tonb;PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
42 d3dhxa1
not modelled
11.8
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: NIL domain-like43 c1bl1A_
not modelled
11.7
38
PDB header: hormone receptorChain: A: PDB Molecule: parathyroid hormone receptor;PDBTitle: pth receptor n-terminus fragment, nmr, 1 structure
44 c2c5sA_
not modelled
11.5
10
PDB header: rna-binding proteinChain: A: PDB Molecule: probable thiamine biosynthesis protein thii;PDBTitle: crystal structure of bacillus anthracis thii, a trna-2 modifying enzyme containing the predicted rna-binding3 thump domain
45 c3echC_
not modelled
11.1
55
PDB header: transcription, transcription regulationChain: C: PDB Molecule: 25-mer fragment of protein armr;PDBTitle: the marr-family repressor mexr in complex with its antirepressor armr
46 d1rtya_
not modelled
10.7
10
Fold: Ferritin-likeSuperfamily: Cobalamin adenosyltransferase-likeFamily: Cobalamin adenosyltransferase47 d1r9da_
not modelled
10.6
15
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like48 d2gskb1
not modelled
10.4
4
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB49 c2jynA_
not modelled
10.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0368 protein ypl225w;PDBTitle: a novel solution nmr structure of protein yst0336 from2 saccharomyces cerevisiae. northeast structural genomics3 consortium target yt51/ontario centre for structural4 proteomics target yst0336
50 d1es6a2
not modelled
10.0
55
Fold: EV matrix proteinSuperfamily: EV matrix proteinFamily: EV matrix protein51 c1es6A_
not modelled
9.8
55
PDB header: viral proteinChain: A: PDB Molecule: matrix protein vp40;PDBTitle: crystal structure of the matrix protein of ebola virus
52 c1xx3A_
not modelled
9.7
3
PDB header: transport proteinChain: A: PDB Molecule: tonb protein;PDBTitle: solution structure of escherichia coli tonb-ctd
53 d1gxma_
not modelled
9.5
25
Fold: alpha/alpha toroidSuperfamily: Family 10 polysaccharide lyaseFamily: Family 10 polysaccharide lyase54 d1n62c1
not modelled
9.0
9
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like55 c3k1dA_
not modelled
8.6
23
PDB header: transferaseChain: A: PDB Molecule: 1,4-alpha-glucan-branching enzyme;PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
56 d1d7qa_
not modelled
8.6
5
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like57 d1x2ga1
not modelled
8.5
19
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SP1160 C-terminal domain-like58 d1yeya2
not modelled
8.1
14
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like59 d1jt8a_
not modelled
7.3
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like60 c2fq2A_
not modelled
7.3
12
PDB header: lipid transportChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of minor conformation of holo-acyl2 carrier protein from malaria parasite plasmodium falciparum
61 d2oeza1
not modelled
7.3
8
Fold: YacF-likeSuperfamily: YacF-likeFamily: YacF-like62 d1h4ra3
not modelled
7.2
11
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM63 c2kw0A_
not modelled
7.0
12
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
64 d3ceda1
not modelled
7.0
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: NIL domain-like65 c2zhzC_
not modelled
7.0
8
PDB header: transferaseChain: C: PDB Molecule: atp:cob(i)alamin adenosyltransferase, putative;PDBTitle: crystal structure of a pduo-type atp:cobalamin adenosyltransferase2 from burkholderia thailandensis
66 c2l9fA_
not modelled
7.0
9
PDB header: transferaseChain: A: PDB Molecule: cale8;PDBTitle: nmr solution structure of meacp
67 d1vi7a2
not modelled
6.9
12
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: YigZ C-terminal domain-like68 c1vi7A_
not modelled
6.8
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yigz;PDBTitle: crystal structure of an hypothetical protein
69 c2hl7A_
not modelled
6.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
70 d1jroa3
not modelled
6.5
17
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like71 c2p4vA_
not modelled
6.5
15
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
72 c2fq8A_
not modelled
6.4
36
PDB header: unknown functionChain: A: PDB Molecule: 2f;PDBTitle: nmr structure of 2f associated with lipid disc
73 c2fq5A_
not modelled
6.4
36
PDB header: unknown functionChain: A: PDB Molecule: peptide 2f;PDBTitle: nmr structure of 2f associated with lipid disc
74 d7mdha2
not modelled
6.3
0
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain75 c1hucC_
not modelled
6.3
18
PDB header: thiol proteaseChain: C: PDB Molecule: cathepsin b;PDBTitle: the refined 2.15 angstroms x-ray crystal structure of human2 liver cathepsin b: the structural basis for its specificity
76 c3sc0A_
not modelled
6.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: methylmalonic aciduria and homocystinuria type c protein;PDBTitle: crystal structure of mmachc (1-238), a human b12 processing enzyme,2 complexed with methylcobalamin
77 d1v8ca2
not modelled
6.2
20
Fold: TBP-likeSuperfamily: MoaD-related protein, C-terminal domainFamily: MoaD-related protein, C-terminal domain78 d2up1a2
not modelled
6.1
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD79 c2ochA_
not modelled
6.1
38
PDB header: chaperoneChain: A: PDB Molecule: hypothetical protein dnj-12;PDBTitle: j-domain of dnj-12 from caenorhabditis elegans
80 c2vgpD_
not modelled
6.1
28
PDB header: transferaseChain: D: PDB Molecule: inner centromere protein a;PDBTitle: crystal structure of aurora b kinase in complex with a2 aminothiazole inhibitor
81 d1uptd_
not modelled
6.1
31
Fold: GRIP domainSuperfamily: GRIP domainFamily: GRIP domain82 d2jeka1
not modelled
6.0
19
Fold: Rv1873-likeSuperfamily: Rv1873-likeFamily: Rv1873-like83 d2r4qa1
not modelled
6.0
12
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like84 c2e5aA_
not modelled
5.9
8
PDB header: ligaseChain: A: PDB Molecule: lipoyltransferase 1;PDBTitle: crystal structure of bovine lipoyltransferase in complex2 with lipoyl-amp
85 d1t3qc1
not modelled
5.9
8
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: CO dehydrogenase flavoprotein C-terminal domain-likeFamily: CO dehydrogenase flavoprotein C-terminal domain-like86 c2l4bA_
not modelled
5.8
8
PDB header: transferaseChain: A: PDB Molecule: acyl carrier protein;PDBTitle: solution structure of a putative acyl carrier protein from anaplasma2 phagocytophilum. seattle structural genomics center for infectious3 disease target anpha.01018.a
87 c2kciA_
not modelled
5.8
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative acyl carrier protein;PDBTitle: solution nmr structure of gmet_2339 from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr141
88 d2q22a1
not modelled
5.8
13
Fold: Ava3019-likeSuperfamily: Ava3019-likeFamily: Ava3019-like89 c2pt7G_
not modelled
5.7
13
PDB header: hydrolase/protein bindingChain: G: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of cag virb11 (hp0525) and an inhibitory protein2 (hp1451)
90 d1vcta1
not modelled
5.7
10
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like91 d2qlvb1
not modelled
5.7
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like92 d1r6ta2
not modelled
5.6
23
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain93 c2k9kA_
not modelled
5.6
17
PDB header: metal transportChain: A: PDB Molecule: tonb2;PDBTitle: molecular characterization of the tonb2 protein from vibrio2 anguillarum
94 d2f15a1
not modelled
5.6
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like95 c3g59A_
not modelled
5.5
27
PDB header: transferaseChain: A: PDB Molecule: fmn adenylyltransferase;PDBTitle: crystal structure of candida glabrata fmn2 adenylyltransferase in complex with atp
96 c2or0B_
not modelled
5.5
7
PDB header: oxidoreductaseChain: B: PDB Molecule: hydroxylase;PDBTitle: structural genomics, the crystal structure of a putative hydroxylase2 from rhodococcus sp. rha1
97 c2dn9A_
not modelled
5.5
21
PDB header: apoptosis, chaperoneChain: A: PDB Molecule: dnaj homolog subfamily a member 3;PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
98 c3oakC_
not modelled
5.5
57
PDB header: transcriptionChain: C: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of a spn1 (iws1)-spt6 complex
99 c3fq9A_
not modelled
5.4
5
PDB header: hormoneChain: A: PDB Molecule: insulin;PDBTitle: design of an insulin analog with enhanced receptor-binding2 selectivity. rationale, structure, and therapeutic3 implications