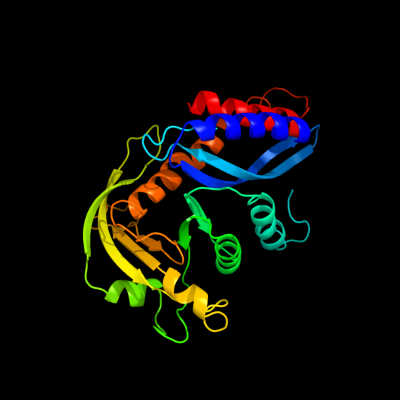
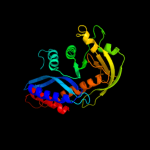
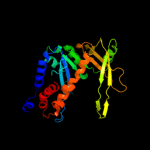
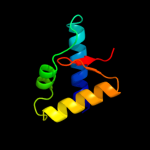
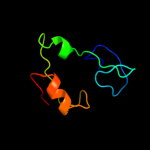
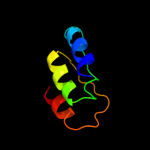
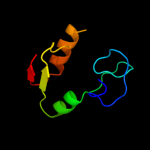
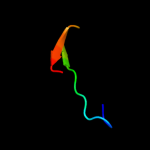
1 d1b7ea_
98.1
11
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)2 d1musa_
98.0
14
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)3 d2v9va2
79.2
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal fragment of elongation factor SelB4 c2v9vA_
66.0
16
PDB header: transcriptionChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: crystal structure of moorella thermoacetica selb(377-511)
5 d2ezha_
52.4
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain6 d2ezia_
49.6
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain7 c3hefB_
39.9
17
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
8 c6paxA_
36.1
27
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeobox protein pax-6;PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
9 c2kzvA_
29.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of cv_0373(175-257) protein from2 chromobacterium violaceum, northeast structural genomics consortium3 target cvr118a
10 c2vp8A_
24.3
19
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase 2;PDBTitle: structure of mycobacterium tuberculosis rv1207
11 d1osna_
20.4
7
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases12 c2plyB_
19.1
16
PDB header: translation/rnaChain: B: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: structure of the mrna binding fragment of elongation factor2 selb in complex with secis rna.
13 c3nswA_
17.5
70
PDB header: immune systemChain: A: PDB Molecule: excretory-secretory protein 2;PDBTitle: crystal structure of ancylostoma ceylanicum excretory-secretory2 protein 2
14 c3a52A_
17.1
15
PDB header: hydrolaseChain: A: PDB Molecule: cold-active alkaline phosphatase;PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
15 d1ajza_
16.1
11
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase16 c3e2dB_
15.9
12
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: the 1.4 a crystal structure of the large and cold-active2 vibrio sp. alkaline phosphatase
17 c3mx7A_
15.3
17
PDB header: apoptosisChain: A: PDB Molecule: fas apoptotic inhibitory molecule 1;PDBTitle: crystal structure analysis of human faim-ntd
18 d3orca_
14.4
21
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors19 c2y5sA_
14.4
17
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase;PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
20 d1pdnc_
13.6
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain21 d1iyxa1
not modelled
13.3
25
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase22 c3r1iB_
not modelled
13.1
25
PDB header: oxidoreductaseChain: B: PDB Molecule: short-chain type dehydrogenase/reductase;PDBTitle: crystal structure of a short-chain type dehydrogenase/reductase from2 mycobacterium marinum
23 c1hj0A_
not modelled
12.2
25
PDB header: actin binding peptideChain: A: PDB Molecule: thymosin beta9;PDBTitle: thymosin beta9
24 d1eyea_
not modelled
11.5
19
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase25 d1dlwa_
not modelled
10.5
13
Fold: Globin-likeSuperfamily: Globin-likeFamily: Truncated hemoglobin26 c2iucB_
not modelled
10.0
23
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structure of alkaline phosphatase from the antarctic2 bacterium tab5
27 d1uuza_
not modelled
9.7
15
Fold: Inhibitor of vertebrate lysozyme, IvySuperfamily: Inhibitor of vertebrate lysozyme, IvyFamily: Inhibitor of vertebrate lysozyme, Ivy28 d1d1la_
not modelled
9.6
24
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors29 d2hxja1
not modelled
9.5
14
Fold: FinO-likeSuperfamily: FinO-likeFamily: FinO-like30 d2gm3a1
not modelled
9.5
13
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like31 c3rf7A_
not modelled
9.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: iron-containing alcohol dehydrogenase;PDBTitle: crystal structure of an iron-containing alcohol dehydrogenase2 (sden_2133) from shewanella denitrificans os-217 at 2.12 a resolution
32 c3emkA_
not modelled
9.1
21
PDB header: oxidoreductaseChain: A: PDB Molecule: glucose/ribitol dehydrogenase;PDBTitle: 2.5a crystal structure of glucose/ribitol dehydrogenase2 from brucella melitensis
33 d2a22a1
not modelled
9.1
20
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: YfcE-like34 d2fgga1
not modelled
9.0
31
Fold: dsRBD-likeSuperfamily: Rv2632c-likeFamily: Rv2632c-like35 c2kvcA_
not modelled
8.8
27
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
36 d1mpga1
not modelled
8.8
8
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: DNA repair glycosylase, 2 C-terminal domains37 c3douA_
not modelled
8.8
20
PDB header: transferaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase j;PDBTitle: crystal structure of methyltransferase involved in cell2 division from thermoplasma volcanicum gss1
38 c2pijB_
not modelled
8.6
16
PDB header: transcriptionChain: B: PDB Molecule: prophage pfl 6 cro;PDBTitle: structure of the cro protein from prophage pfl 6 in pseudomonas2 fluorescens pf-5
39 d4croa_
not modelled
8.3
24
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors40 c3pk0B_
not modelled
8.3
12
PDB header: oxidoreductaseChain: B: PDB Molecule: short-chain dehydrogenase/reductase sdr;PDBTitle: crystal structure of short-chain dehydrogenase/reductase sdr from2 mycobacterium smegmatis
41 d1e8qa_
not modelled
8.2
13
Fold: Cellulose docking domain, dockeringSuperfamily: Cellulose docking domain, dockeringFamily: Cellulose docking domain, dockering42 d2ezla_
not modelled
8.1
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain43 d1vi7a1
not modelled
8.0
19
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: YigZ N-terminal domain-like44 d1xb4a2
not modelled
7.8
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain45 c2rfpA_
not modelled
7.8
10
PDB header: hydrolaseChain: A: PDB Molecule: putative ntp pyrophosphohydrolase;PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
46 c3gk0H_
not modelled
7.2
24
PDB header: transferaseChain: H: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
47 c2z1nA_
not modelled
7.1
24
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydrogenase;PDBTitle: crystal structure of ape0912 from aeropyrum pernix k1
48 d3bpya1
not modelled
7.0
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain49 d1s69a_
not modelled
7.0
20
Fold: Globin-likeSuperfamily: Globin-likeFamily: Truncated hemoglobin50 c2hxjF_
not modelled
7.0
14
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a protein of unknown function nmb1681 from2 neisseria meningitidis mc58, possible nucleic acid binding protein
51 d1cyda_
not modelled
7.0
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases52 d2c6ya1
not modelled
6.8
3
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain53 d2ptza1
not modelled
6.8
20
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase54 c3l1pA_
not modelled
6.7
17
PDB header: transcription/dnaChain: A: PDB Molecule: pou domain, class 5, transcription factor 1;PDBTitle: pou protein:dna complex
55 c1c94B_
not modelled
6.7
38
PDB header: gene regulationChain: B: PDB Molecule: retro-gcn4 leucine zipper;PDBTitle: reversing the sequence of the gcn4 leucine zipper does not2 affect its fold.
56 c3aq8A_
not modelled
6.5
13
PDB header: oxygen bindingChain: A: PDB Molecule: group 1 truncated hemoglobin;PDBTitle: crystal structure of truncated hemoglobin from tetrahymena pyriformis,2 q46e mutant, fe(iii) form
57 c3ol4B_
not modelled
6.5
35
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
58 c3ak4C_
not modelled
6.4
13
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-dependent quinuclidinone reductase;PDBTitle: crystal structure of nadh-dependent quinuclidinone reductase from2 agrobacterium tumefaciens
59 c2wdzD_
not modelled
6.4
18
PDB header: oxidoreductaseChain: D: PDB Molecule: short-chain dehydrogenase/reductase;PDBTitle: crystal structure of the short chain dehydrogenase2 galactitol-dehydrogenase (gatdh) of rhodobacter3 sphaeroides in complex with nad+ and 1,2-pentandiol
60 d2czda1
not modelled
6.3
22
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase61 c1e17A_
not modelled
6.3
7
PDB header: dna binding domainChain: A: PDB Molecule: afx;PDBTitle: solution structure of the dna binding domain of the human2 forkhead transcription factor afx (foxo4)
62 c3cxtA_
not modelled
6.2
24
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydrogenase with different specificities;PDBTitle: quaternary complex structure of gluconate 5-dehydrogenase from2 streptococcus suis type 2
63 d1w6ta1
not modelled
6.2
24
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase64 c3b9nB_
not modelled
6.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: alkane monoxygenase;PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
65 c2kl8A_
not modelled
6.0
22
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
66 d1l1ya_
not modelled
6.0
57
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain67 c1l2aD_
not modelled
6.0
57
PDB header: hydrolaseChain: D: PDB Molecule: cellobiohydrolase;PDBTitle: the crystal structure and catalytic mechanism of2 cellobiohydrolase cels, the major enzymatic component of3 the clostridium thermocellum cellulosome
68 c1qcrD_
not modelled
5.9
6
PDB header: PDB COMPND: 69 d1g31a_
not modelled
5.8
20
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES70 c3gdfA_
not modelled
5.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nadp-dependent mannitol dehydrogenase;PDBTitle: crystal structure of the nadp-dependent mannitol dehydrogenase from2 cladosporium herbarum.
71 d2abka_
not modelled
5.4
13
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: Endonuclease III72 c2o2sA_
not modelled
5.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: enoyl-acyl carrier reductase;PDBTitle: the structure of t. gondii enoyl acyl carrier protein reductase in2 complex with nad and triclosan
73 d1m5wa_
not modelled
5.4
24
Fold: TIM beta/alpha-barrelSuperfamily: Pyridoxine 5'-phosphate synthaseFamily: Pyridoxine 5'-phosphate synthase74 c3cwbQ_
not modelled
5.3
5
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
75 c1htrP_
not modelled
5.3
20
PDB header: aspartyl proteaseChain: P: PDB Molecule: progastricsin (pro segment);PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
76 d1un2a_
not modelled
5.2
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like77 c3co7C_
not modelled
5.2
11
PDB header: transcription/dnaChain: C: PDB Molecule: forkhead box protein o1;PDBTitle: crystal structure of foxo1 dbd bound to dbe2 dna
78 d2giab1
not modelled
5.2
50
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: Guide RNA binding protein gBP79 c2giaB_
not modelled
5.2
50
PDB header: translationChain: B: PDB Molecule: mitochondrial rna-binding protein 1;PDBTitle: crystal structures of trypanosoma bruciei mrp1/mrp2
80 d2i7pa1
not modelled
5.2
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like81 c3gafF_
not modelled
5.1
15
PDB header: oxidoreductaseChain: F: PDB Molecule: 7-alpha-hydroxysteroid dehydrogenase;PDBTitle: 2.2a crystal structure of 7-alpha-hydroxysteroid2 dehydrogenase from brucella melitensis
82 d2c4ka1
not modelled
5.1
23
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like83 c3hluA_
not modelled
5.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein duf2179;PDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 duf2179 from eubacterium ventriosum
84 c2bmbA_
not modelled
5.1
20
PDB header: transferaseChain: A: PDB Molecule: folic acid synthesis protein fol1;PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
85 d1gpqa_
not modelled
5.1
16
Fold: Inhibitor of vertebrate lysozyme, IvySuperfamily: Inhibitor of vertebrate lysozyme, IvyFamily: Inhibitor of vertebrate lysozyme, Ivy86 d2o23a1
not modelled
5.1
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases87 c2x4mD_
not modelled
5.1
27
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
88 d1g9ga_
not modelled
5.0
26
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain