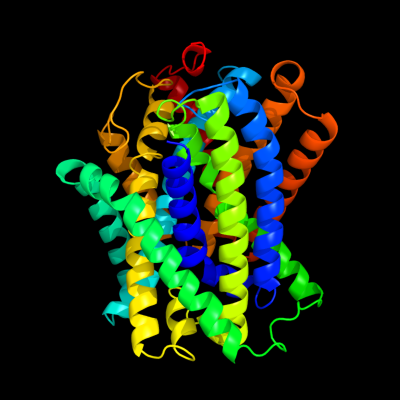
1 c3giaA_
98.0
14
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
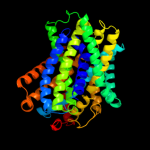
2 c2xq2A_
97.4
16
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
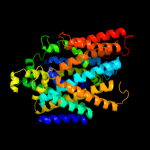
3 c2jlnA_
97.2
14
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
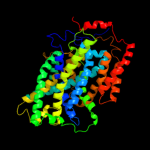
4 c3dh4A_
88.3
15
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
5 c3lrcC_
82.6
13
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
6 c3a8qB_
31.6
70
PDB header: signaling proteinChain: B: PDB Molecule: t-lymphoma invasion and metastasis-inducingPDBTitle: low-resolution crystal structure of the tiam2 phccex domain
7 c3a8nA_
29.7
58
PDB header: signaling proteinChain: A: PDB Molecule: t-lymphoma invasion and metastasis-inducingPDBTitle: crystal structure of the tiam1 phccex domain
8 c3pl0B_
21.5
33
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a bsma homolog (mpe_a2762) from methylobium2 petroleophilum pm1 at 1.91 a resolution
9 d1rp4a_
17.7
41
Fold: ERO1-likeSuperfamily: ERO1-likeFamily: ERO1-like10 c3hd6A_
16.3
11
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: ammonium transporter rh type c;PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
11 c2jp3A_
14.1
16
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
12 c3b9yA_
12.3
14
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
13 c2k9pA_
12.3
24
PDB header: membrane proteinChain: A: PDB Molecule: pheromone alpha factor receptor;PDBTitle: structure of tm1_tm2 in lppg micelles
14 d2axtj1
11.5
21
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like15 d2bfdb2
11.4
8
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain16 c3ahrA_
10.6
56
PDB header: oxidoreductaseChain: A: PDB Molecule: ero1-like protein alpha;PDBTitle: inactive human ero1
17 d2awia1
10.5
41
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: PrgX N-terminal domain-like18 c3chxE_
9.7
40
PDB header: membrane proteinChain: E: PDB Molecule: pmob;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
19 d1qs0b2
9.2
19
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain20 d1h5pa_
9.2
27
Fold: SAND domain-likeSuperfamily: SAND domain-likeFamily: SAND domain21 d1oqja_
not modelled
9.2
27
Fold: SAND domain-likeSuperfamily: SAND domain-likeFamily: SAND domain22 d1afra_
not modelled
9.1
30
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like23 d2fug61
not modelled
9.0
31
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nq06-like24 d1s6la1
not modelled
8.5
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like25 d1ufna_
not modelled
8.4
27
Fold: SAND domain-likeSuperfamily: SAND domain-likeFamily: SAND domain26 c2ktlA_
not modelled
8.1
16
PDB header: ligaseChain: A: PDB Molecule: tyrosyl-trna synthetase;PDBTitle: structure of c-terminal domain from mttyrrs of a. nidulans
27 d2etja1
not modelled
8.0
67
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H28 c2etjA_
not modelled
8.0
67
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease hii;PDBTitle: crystal structure of ribonuclease hii (ec 3.1.26.4) (rnase hii)2 (tm0915) from thermotoga maritima at 1.74 a resolution
29 c2jo1A_
not modelled
7.8
8
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
30 c3iymA_
not modelled
7.8
35
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: backbone trace of the capsid protein dimer of a fungal partitivirus2 from electron cryomicroscopy and homology modeling
31 d1ik6a2
not modelled
7.5
19
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain32 d2ozlb2
not modelled
7.0
14
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain33 d5csma_
not modelled
6.9
16
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Allosteric chorismate mutase34 c3pppA_
not modelled
6.5
11
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine/carnitine/choline-binding protein;PDBTitle: structures of the substrate-binding protein provide insights into the2 multiple compatible solutes binding specificities of bacillus3 subtilis abc transporter opuc
35 c1i3aA_
not modelled
6.3
56
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease hii;PDBTitle: rnase hii from archaeoglobus fulgidus with cobalt hexammine2 chloride
36 d1i39a_
not modelled
6.3
56
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H37 d1ekea_
not modelled
6.3
67
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H38 c3kioA_
not modelled
5.9
67
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease h2 subunit a;PDBTitle: mouse rnase h2 complex
39 c2qx5B_
not modelled
5.8
12
PDB header: transport proteinChain: B: PDB Molecule: nucleoporin nic96;PDBTitle: structure of nucleoporin nic96
40 d1uaxa_
not modelled
5.8
67
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H41 c3zy6A_
not modelled
5.8
20
PDB header: transferaseChain: A: PDB Molecule: putative gdp-fucose protein o-fucosyltransferase 1;PDBTitle: crystal structure of pofut1 in complex with gdp-fucose2 (crystal-form-ii)
42 c2k42A_
not modelled
5.7
15
PDB header: signaling proteinChain: A: PDB Molecule: wiskott-aldrich syndrome protein;PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
43 d1w85b2
not modelled
5.6
34
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain44 c2d0bA_
not modelled
5.5
44
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease hiii;PDBTitle: crystal structure of bst-rnase hiii in complex with mg2+
45 d1io2a_
not modelled
5.5
67
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H46 d1llda2
not modelled
5.4
29
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain47 d1i0za2
not modelled
5.4
25
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain48 c2elnA_
not modelled
5.4
38
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 11th c2h2 zinc finger of human2 zinc finger protein 406
49 d9ldta2
not modelled
5.4
25
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain50 d2yrka1
not modelled
5.3
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger51 c2lbfA_
not modelled
5.3
43
PDB header: ribosomal proteinChain: A: PDB Molecule: 60s acidic ribosomal protein p1;PDBTitle: solution structure of the dimerization domain of human ribosomal2 protein p1/p2 heterodimer
52 d1nekd_
not modelled
5.3
11
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)