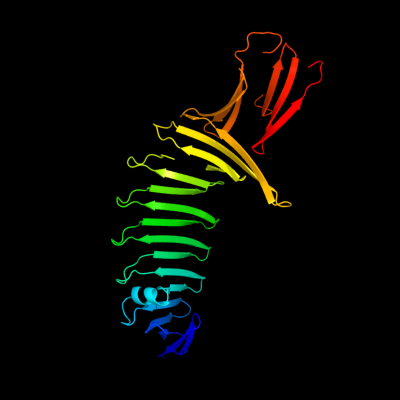
1 c2oy7A_
95.2
16
PDB header: membrane proteinChain: A: PDB Molecule: outer surface protein a;PDBTitle: the crystal structure of ospa mutant
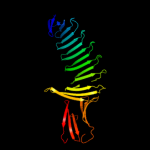
2 d1x3za1
52.2
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core3 d1ulva3
45.0
19
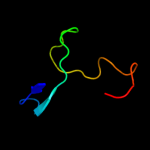
Fold: Immunoglobulin-like beta-sandwichSuperfamily: CBD9-likeFamily: Glucodextranase, domain C4 c1n7dA_
44.1
11
PDB header: lipid transportChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: extracellular domain of the ldl receptor
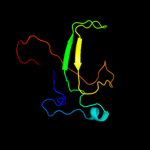
5 c3eswA_
31.3
19
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
6 c3mswA_
30.2
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (bf3112) from2 bacteroides fragilis nctc 9343 at 1.90 a resolution
7 d1oeya_
29.6
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain8 d2f4ma1
29.4
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core9 c2ivzD_
26.0
11
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: protein tolb;PDBTitle: structure of tolb in complex with a peptide of the colicin2 e9 t-domain
10 c3rwxA_
24.5
21
PDB header: transport proteinChain: A: PDB Molecule: hypothetical bacterial outer membrane protein;PDBTitle: crystal structure of a hypothetical bacterial outer membrane protein2 (bf2706) from bacteroides fragilis nctc 9343 at 2.40 a resolution
11 c2w8bB_
23.3
12
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
12 c1mjeA_
23.3
28
PDB header: gene regulation/antitumor protein/dnaChain: A: PDB Molecule: breast cancer 2;PDBTitle: structure of a brca2-dss1-ssdna complex
13 c1miuA_
23.0
27
PDB header: gene regulation/antitumor proteinChain: A: PDB Molecule: breast cancer type 2 susceptibility protein;PDBTitle: structure of a brca2-dss1 complex
14 d1khda2
22.8
35
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain15 c2kijA_
22.7
25
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the actuator domain of the copper-2 transporting atpase atp7a
16 c2fz0A_
22.2
19
PDB header: membrane proteinChain: A: PDB Molecule: v-snare component of the vacuolar snare complexPDBTitle: identification of yeast r-snare nyv1p as a novel longin2 domain protein
17 c1j5qB_
21.5
26
PDB header: viral proteinChain: B: PDB Molecule: major capsid protein;PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
18 d1qs0b1
18.8
32
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Branched-chain alpha-keto acid dehydrogenase Pyr module19 d1o5ua_
18.5
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111220 d1bcga_
16.2
35
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Long-chain scorpion toxins21 c2hc8A_
not modelled
15.5
30
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
22 c1iyjB_
not modelled
15.2
27
PDB header: gene regulation/antitumor proteinChain: B: PDB Molecule: breast cancer susceptibility;PDBTitle: structure of a brca2-dss1 complex
23 c3e5zA_
not modelled
14.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative gluconolactonase;PDBTitle: x-ray structure of the putative gluconolactonase in protein family2 pf08450. northeast structural genomics consortium target drr130.
24 d1pk6c_
not modelled
14.0
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like25 d1ei5a1
not modelled
13.9
32
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains26 c1dskA_
not modelled
13.6
100
PDB header: viral peptideChain: A: PDB Molecule: vpr protein;PDBTitle: nmr solution structure of vpr59_86, 20 structures
27 c2kz5A_
not modelled
13.5
18
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
28 d1miua3
not modelled
13.4
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB29 c2x12A_
not modelled
13.2
23
PDB header: cell adhesionChain: A: PDB Molecule: fimbriae-associated protein fap1;PDBTitle: ph-induced modulation of streptococcus parasanguinis2 adhesion by fap1 fimbriae
30 d1bcoa1
not modelled
13.2
17
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain31 d1vk8a_
not modelled
13.1
14
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like32 c2ka3C_
not modelled
13.0
36
PDB header: structural proteinChain: C: PDB Molecule: emilin-1;PDBTitle: structure of emilin-1 c1q-like domain
33 d1pzqa_
not modelled
12.6
32
Fold: Dimerisation interlockSuperfamily: Docking domain A of the erythromycin polyketide synthase (DEBS)Family: Docking domain A of the erythromycin polyketide synthase (DEBS)34 d1pk6b_
not modelled
12.5
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like35 d1iyjb3
not modelled
12.4
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB36 c2cseU_
not modelled
12.2
19
PDB header: virusChain: U: PDB Molecule: guanylyltransferase;PDBTitle: features of reovirus outer-capsid protein mu1 revealed by2 electron and image reconstruction of the virion at 7.0-a3 resolution
37 d2joya1
not modelled
11.9
7
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e38 d1brwa3
not modelled
11.6
26
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain39 d1lxna_
not modelled
11.5
15
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like40 c3bmxB_
not modelled
11.5
25
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
41 d1c3ha_
not modelled
11.3
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like42 c3dczA_
not modelled
11.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: putative rnfg subunit of electron transport complex;PDBTitle: crystal structure of a putative rnfg subunit of electron transport2 complex (tm0246) from thermotoga maritima at 1.65 a resolution
43 d1m3ya2
not modelled
11.0
33
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group II dsDNA viruses VPFamily: Major capsid protein vp5444 d1uerc2
not modelled
10.9
20
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain45 c1zrxA_
not modelled
10.9
23
PDB header: antimicrobial protein, antibioticChain: A: PDB Molecule: stomoxyn;PDBTitle: solution structure of stomoxyn in h20/tfe 50%
46 c3fnkA_
not modelled
10.9
23
PDB header: structural proteinChain: A: PDB Molecule: cellulosomal scaffoldin adaptor protein b;PDBTitle: crystal structure of the second type ii cohesin module from2 the cellulosomal adaptor scaa scaffoldin of acetivibrio3 cellulolyticus
47 d2tpta3
not modelled
10.7
26
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain48 c3izcN_
not modelled
10.6
21
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
49 c2jpnA_
not modelled
10.5
26
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase uvsw;PDBTitle: solution structure of t4 bacteriophage helicase uvsw.1
50 c3h1sB_
not modelled
10.2
15
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of superoxide dismutase from francisella tularensis2 subsp. tularensis schu s4
51 c3iz5N_
not modelled
10.2
13
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
52 c1m4xC_
not modelled
10.0
33
PDB header: virusChain: C: PDB Molecule: pbcv-1 virus capsid;PDBTitle: pbcv-1 virus capsid, quasi-atomic model
53 c1trlB_
not modelled
10.0
19
PDB header: hydrolase (metalloprotease)Chain: B: PDB Molecule: thermolysin fragment 255 - 316;PDBTitle: nmr solution structure of the c-terminal fragment 255-3162 of thermolysin: a dimer formed by subunits having the3 native structure
54 d2pyta1
not modelled
10.0
15
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like55 c2oajA_
not modelled
10.0
12
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: protein sni1;PDBTitle: crystal structure of sro7 from s. cerevisiae
56 c2jttD_
not modelled
9.9
36
PDB header: calcium binding protein/antitumor proteiChain: D: PDB Molecule: calcyclin-binding protein;PDBTitle: solution structure of calcium loaded s100a6 bound to c-2 terminal siah-1 interacting protein
57 d1lxja_
not modelled
9.5
12
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like58 c3lk6A_
not modelled
9.3
25
PDB header: hydrolaseChain: A: PDB Molecule: lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
59 c3jroA_
not modelled
9.1
13
PDB header: transport protein, structural proteinChain: A: PDB Molecule: fusion protein of protein transport protein sec13PDBTitle: nup84-nup145c-sec13 edge element of the npc lattice
60 c2gacA_
not modelled
9.1
13
PDB header: hydrolaseChain: A: PDB Molecule: glycosylasparaginase;PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
61 d1sknp_
not modelled
9.0
11
Fold: A DNA-binding domain in eukaryotic transcription factorsSuperfamily: A DNA-binding domain in eukaryotic transcription factorsFamily: A DNA-binding domain in eukaryotic transcription factors62 d1ejxc1
not modelled
9.0
26
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: alpha-Subunit of urease63 d1dfma_
not modelled
8.9
47
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BglII64 c2jtmA_
not modelled
8.9
29
PDB header: dna binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of sso6901 from sulfolobus solfataricus2 p2
65 d1jnpa_
not modelled
8.7
24
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products66 d1a1xa_
not modelled
8.7
19
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products67 c3nv4A_
not modelled
8.7
28
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin 9 short isoform variant;PDBTitle: crystal structure of human galectin-9 c-terminal crd in complex with2 sialyllactose
68 c2ksnA_
not modelled
8.6
18
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin domain-containing protein 2;PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
69 d1pk6a_
not modelled
8.6
22
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like70 c1bdeA_
not modelled
8.4
27
PDB header: aidsChain: A: PDB Molecule: vpr protein;PDBTitle: helical structure of polypeptides from the c-terminal half2 of hiv-1 vpr, nmr, 20 structures
71 c3nqhA_
not modelled
8.4
15
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: crystal structure of a glycosyl hydrolase (bt_2959) from bacteroides2 thetaiotaomicron vpi-5482 at 2.11 a resolution
72 d2f1fa1
not modelled
8.3
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like73 d1f00i2
not modelled
8.3
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments74 d1xata_
not modelled
8.3
21
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like75 c2vtwF_
not modelled
8.2
32
PDB header: viral proteinChain: F: PDB Molecule: fiber protein 2;PDBTitle: structure of the c-terminal head domain of the fowl2 adenovirus type 1 short fibre
76 c1gv3B_
not modelled
8.2
11
PDB header: manganese superoxide dismutaseChain: B: PDB Molecule: manganese superoxide dismutase;PDBTitle: the 2.0 angstrom resolution structure of the catalytic2 portion of a cyanobacterial membrane-bound manganese3 superoxide dismutase
77 c1dfwA_
not modelled
8.2
57
PDB header: immune systemChain: A: PDB Molecule: lung surfactant protein b;PDBTitle: conformational mapping of the n-terminal segment of2 surfactant protein b in lipid using 13c-enhanced fourier3 transform infrared spectroscopy (ftir)
78 d1x6va1
not modelled
8.2
19
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ATP sulfurylase N-terminal domain79 d2pc6a2
not modelled
8.1
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like80 c2wlgA_
not modelled
7.9
26
PDB header: transferaseChain: A: PDB Molecule: polysialic acid o-acetyltransferase;PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
81 d1e9yb1
not modelled
7.9
26
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: alpha-Subunit of urease82 d1o91a_
not modelled
7.9
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like83 c1o91B_
not modelled
7.9
31
PDB header: collagenChain: B: PDB Molecule: collagen alpha 1(viii) chain;PDBTitle: crystal structure of a collagen viii nc1 domain trimer
84 c2iu9C_
not modelled
7.8
23
PDB header: transferaseChain: C: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosaminePDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
85 d2q09a1
not modelled
7.8
80
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Imidazolonepropionase-like86 d2jdqd1
not modelled
7.7
20
Fold: PB2 C-terminal domain-likeSuperfamily: PB2 C-terminal domain-likeFamily: PB2 C-terminal domain-like87 d2i5ua1
not modelled
7.7
17
Fold: DnaD domain-likeSuperfamily: DnaD domain-likeFamily: DnaD domain88 d1w9pa2
not modelled
7.6
18
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain89 d1jsga_
not modelled
7.5
24
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products90 d2hqya2
not modelled
7.5
19
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: FemXAB nonribosomal peptidyltransferases91 d1azpa_
not modelled
7.5
17
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea92 c2a03A_
not modelled
7.5
17
PDB header: oxidoreductaseChain: A: PDB Molecule: fe-superoxide dismutase homolog;PDBTitle: superoxide dismutase protein from plasmodium berghei
93 c3cl5A_
not modelled
7.4
20
PDB header: hydrolaseChain: A: PDB Molecule: hemagglutinin-esterase;PDBTitle: structure of coronavirus hemagglutinin-esterase in complex with 4,9-o-2 diacetyl sialic acid
94 d1r7aa1
not modelled
7.4
36
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain95 d1thqa_
not modelled
7.4
50
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane enzyme PagP96 d1zaka2
not modelled
7.4
33
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain97 c2kjmA_
not modelled
7.4
16
PDB header: rna binding proteinChain: A: PDB Molecule: histone rna hairpin-binding protein;PDBTitle: solution structure of slbp rna binding domain fragment
98 c1t3dB_
not modelled
7.3
30
PDB header: transferaseChain: B: PDB Molecule: serine acetyltransferase;PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
99 d1s6la1
not modelled
7.3
44
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like