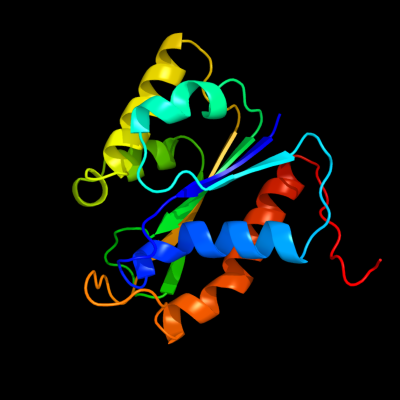
| 1 | d1cfza_
|
|
 |
100.0 |
38 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Hydrogenase maturating endopeptidase HybD |
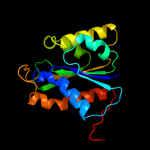
| 2 | c2e85B_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase 3 maturation protease;
PDBTitle: crystal structure of the hydrogenase 3 maturation protease
|
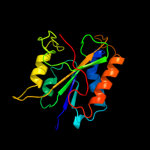
| 3 | c3pu6A_
|
|
 |
100.0 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of an uncharacterized protein from wolinella2 succinogenes
|
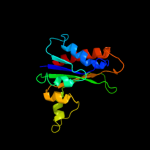
| 4 | d1c8ba_
|
|
 |
96.9 |
28 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Germination protease |
| 5 | c3fvwA_
|
|
 |
90.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative nad(p)h-dependent fmn reductase;
PDBTitle: crystal structure of the q8dwd8_strmu protein from2 streptococcus mutans. northeast structural genomics3 consortium target smr99.
|
| 6 | d9ldta1
|
|
 |
85.1 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 7 | c2z2jA_
|
|
 |
84.2 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from mycobacterium2 tuberculosis
|
| 8 | c2cukC_
|
|
 |
83.4 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
| 9 | d1fmfa_
|
|
 |
81.7 |
11 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 10 | c2ph5A_
|
|
 |
80.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
| 11 | d1a9xa3
|
|
 |
79.8 |
16 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 12 | d2ldxa1
|
|
 |
78.9 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 13 | c2vhyB_
|
|
 |
78.2 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of apo l-alanine dehydrogenase from2 mycobacterium tuberculosis
|
| 14 | d1i0za1
|
|
 |
77.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 15 | d1jf8a_
|
|
 |
77.2 |
15 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 16 | d5pnta_
|
|
 |
76.9 |
19 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 17 | d5ldha1
|
|
 |
76.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 18 | d1dg9a_
|
|
 |
76.0 |
19 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 19 | d2c42a3
|
|
 |
75.4 |
15 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Pyruvate-ferredoxin oxidoreductase, PFOR, domain II |
| 20 | d1i10a1
|
|
 |
74.1 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 21 | c3d0oA_ |
|
not modelled |
73.3 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase 1;
PDBTitle: crystal structure of lactate dehydrogenase from2 staphylococcus aureus
|
| 22 | d1pjca1 |
|
not modelled |
72.5 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 23 | d2gv8a1 |
|
not modelled |
69.0 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 24 | c3ggpA_ |
|
not modelled |
68.6 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
|
| 25 | c4a26B_ |
|
not modelled |
67.1 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
| 26 | c1wwkA_ |
|
not modelled |
67.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 27 | c3b1fA_ |
|
not modelled |
66.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 mutans
|
| 28 | d2jfga1 |
|
not modelled |
66.8 |
24 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 29 | c3gucB_ |
|
not modelled |
66.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:ubiquitin-like modifier-activating enzyme 5;
PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
|
| 30 | d2dlda1 |
|
not modelled |
64.8 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 31 | c2gi4A_ |
|
not modelled |
64.6 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:possible phosphotyrosine protein phosphatase;
PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
|
| 32 | d1gdha1 |
|
not modelled |
64.5 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 33 | c1d4fD_ |
|
not modelled |
63.6 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
| 34 | d1qp8a1 |
|
not modelled |
62.6 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 35 | c3dapB_ |
|
not modelled |
62.0 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:diaminopimelic acid dehydrogenase;
PDBTitle: c. glutamicum dap dehydrogenase in complex with nadp+ and2 the inhibitor 5s-isoxazoline
|
| 36 | c2rirA_ |
|
not modelled |
60.7 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 37 | c2yxbA_ |
|
not modelled |
60.6 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 38 | d1wu2a3 |
|
not modelled |
60.4 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 39 | d1np3a2 |
|
not modelled |
60.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 40 | c1gdhA_ |
|
not modelled |
60.0 |
17 |
PDB header:oxidoreductase(choh (d)-nad(p)+ (a))
Chain: A: PDB Molecule:d-glycerate dehydrogenase;
PDBTitle: crystal structure of a nad-dependent d-glycerate2 dehydrogenase at 2.4 angstroms resolution
|
| 41 | c3tovB_ |
|
not modelled |
59.3 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 9;
PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
|
| 42 | c1ez4B_ |
|
not modelled |
59.0 |
37 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of non-allosteric l-lactate dehydrogenase2 from lactobacillus pentosus at 2.3 angstrom resolution
|
| 43 | d1ygya1 |
|
not modelled |
58.8 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 44 | c3gvxA_ |
|
not modelled |
58.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerate dehydrogenase related protein;
PDBTitle: crystal structure of glycerate dehydrogenase related2 protein from thermoplasma acidophilum
|
| 45 | d1hyha1 |
|
not modelled |
57.7 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 46 | d2ptha_ |
|
not modelled |
57.4 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Peptidyl-tRNA hydrolase-like
Family:Peptidyl-tRNA hydrolase-like |
| 47 | d1n1ea2 |
|
not modelled |
56.1 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 48 | c2g5cD_ |
|
not modelled |
56.1 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
|
| 49 | d1mx3a1 |
|
not modelled |
56.1 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 50 | c3vh1A_ |
|
not modelled |
55.7 |
31 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
|
| 51 | d1f0ya2 |
|
not modelled |
55.3 |
33 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 52 | c1m6vE_ |
|
not modelled |
55.3 |
15 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 53 | c3d64A_ |
|
not modelled |
55.2 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
| 54 | c3d1lB_ |
|
not modelled |
55.2 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
| 55 | c3fi9B_ |
|
not modelled |
55.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of malate dehydrogenase from porphyromonas2 gingivalis
|
| 56 | c3neaA_ |
|
not modelled |
54.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from francisella2 tularensis
|
| 57 | c3v2iA_ |
|
not modelled |
54.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: structure of a peptidyl-trna hydrolase (pth) from burkholderia2 thailandensis
|
| 58 | c2g76A_ |
|
not modelled |
54.2 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 59 | c2dbqA_ |
|
not modelled |
54.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxylate reductase;
PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
|
| 60 | c1np3B_ |
|
not modelled |
54.1 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 61 | c3d4oA_ |
|
not modelled |
54.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 62 | d1a9xa4 |
|
not modelled |
53.8 |
14 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 63 | d2ftsa3 |
|
not modelled |
53.7 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 64 | c2fnzA_ |
|
not modelled |
53.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
|
| 65 | c3h9gA_ |
|
not modelled |
53.4 |
24 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:mccb protein;
PDBTitle: crystal structure of e. coli mccb + mcca-n7isoasn
|
| 66 | d1f06a1 |
|
not modelled |
52.2 |
35 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 67 | d1e5qa1 |
|
not modelled |
52.1 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 68 | c3vh3A_ |
|
not modelled |
51.7 |
17 |
PDB header:metal binding protein/protein transport
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of atg7ctd-atg8 complex
|
| 69 | c2ldxA_ |
|
not modelled |
51.7 |
26 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-lactate dehydrogenase;
PDBTitle: characterization of the antigenic sites on the refined 3-2 angstroms resolution structure of mouse testicular lactate3 dehydrogenase c4
|
| 70 | c3ktdC_ |
|
not modelled |
51.4 |
26 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 71 | d1sc6a1 |
|
not modelled |
50.6 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 72 | c1vqwB_ |
|
not modelled |
50.4 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein with similarity to flavin-containing
PDBTitle: crystal structure of a protein with similarity to flavin-2 containing monooxygenases and to mammalian dimethylalanine3 monooxygenases
|
| 73 | c3k96B_ |
|
not modelled |
50.4 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 74 | d1li4a1 |
|
not modelled |
50.3 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 75 | c2xdoC_ |
|
not modelled |
50.1 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:tetx2 protein;
PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
|
| 76 | d1ez4a1 |
|
not modelled |
49.6 |
34 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 77 | d1j4aa1 |
|
not modelled |
49.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 78 | c3gznB_ |
|
not modelled |
49.3 |
27 |
PDB header:protein binding/ligase
Chain: B: PDB Molecule:nedd8-activating enzyme e1 catalytic subunit;
PDBTitle: structure of nedd8-activating enzyme in complex with nedd82 and mln4924
|
| 79 | c3kboB_ |
|
not modelled |
49.1 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate/hydroxypyruvate reductase a;
PDBTitle: 2.14 angstrom crystal structure of putative oxidoreductase (ycdw) from2 salmonella typhimurium in complex with nadp
|
| 80 | c2eklA_ |
|
not modelled |
48.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|
| 81 | c3n58D_ |
|
not modelled |
48.8 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
| 82 | c2q1uA_ |
|
not modelled |
48.7 |
16 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmf in2 complex with nad+ and udp
|
| 83 | c2cwdA_ |
|
not modelled |
48.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight phosphotyrosine protein phosphatase;
PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
|
| 84 | c3n7uD_ |
|
not modelled |
47.5 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
| 85 | d2g5ca2 |
|
not modelled |
47.1 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 86 | d1ryba_ |
|
not modelled |
46.9 |
23 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Peptidyl-tRNA hydrolase-like
Family:Peptidyl-tRNA hydrolase-like |
| 87 | d1dxya1 |
|
not modelled |
46.7 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 88 | c2uyyD_ |
|
not modelled |
46.7 |
28 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
| 89 | c2x2oA_ |
|
not modelled |
46.6 |
27 |
PDB header:flavoprotein
Chain: A: PDB Molecule:nrdi protein;
PDBTitle: the flavoprotein nrdi from bacillus cereus with the2 initially oxidized fmn cofactor in an intermediate3 radiation reduced state
|
| 90 | c1hyhA_ |
|
not modelled |
46.3 |
31 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
| 91 | d1rlja_ |
|
not modelled |
46.3 |
23 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavoprotein NrdI |
| 92 | c1j4aA_ |
|
not modelled |
45.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: insights into domain closure, substrate specificity and2 catalysis of d-lactate dehydrogenase from lactobacillus3 bulgaricus
|
| 93 | d1uz5a3 |
|
not modelled |
45.3 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 94 | d2vapa1 |
|
not modelled |
44.9 |
27 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 95 | d1p8aa_ |
|
not modelled |
44.6 |
27 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 96 | d2qwxa1 |
|
not modelled |
44.2 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 97 | d1llca1 |
|
not modelled |
43.9 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 98 | c3dzbA_ |
|
not modelled |
43.4 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
|
| 99 | d2hmva1 |
|
not modelled |
43.1 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 100 | d1leha1 |
|
not modelled |
42.5 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 101 | d1b0aa1 |
|
not modelled |
42.4 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 102 | d1t0ia_ |
|
not modelled |
42.2 |
10 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 103 | d1uxja1 |
|
not modelled |
41.8 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 104 | d1a4ia1 |
|
not modelled |
41.6 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 105 | d1v8ba1 |
|
not modelled |
41.6 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 106 | d1hdoa_ |
|
not modelled |
41.6 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 107 | d2bkaa1 |
|
not modelled |
41.0 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 108 | d1d4aa_ |
|
not modelled |
41.0 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 109 | c2g1uA_ |
|
not modelled |
40.8 |
29 |
PDB header:transport protein
Chain: A: PDB Molecule:hypothetical protein tm1088a;
PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
|
| 110 | c3gviB_ |
|
not modelled |
40.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of lactate/malate dehydrogenase from2 brucella melitensis in complex with adp
|
| 111 | c2omeA_ |
|
not modelled |
40.7 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:c-terminal-binding protein 2;
PDBTitle: crystal structure of human ctbp2 dehydrogenase complexed with nad(h)
|
| 112 | d1jl3a_ |
|
not modelled |
40.5 |
13 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 113 | c3nklA_ |
|
not modelled |
40.1 |
26 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
| 114 | c3lcmB_ |
|
not modelled |
40.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
|
| 115 | c2gcgB_ |
|
not modelled |
40.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 116 | c3oetF_ |
|
not modelled |
38.9 |
25 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:erythronate-4-phosphate dehydrogenase;
PDBTitle: d-erythronate-4-phosphate dehydrogenase complexed with nad
|
| 117 | c2j6iC_ |
|
not modelled |
38.7 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:formate dehydrogenase;
PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal2 mutant
|
| 118 | c3jviA_ |
|
not modelled |
38.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase;
PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
|
| 119 | d1fcda1 |
|
not modelled |
38.4 |
29 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 120 | c1cjcA_ |
|
not modelled |
38.3 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (adrenodoxin reductase);
PDBTitle: structure of adrenodoxin reductase of mitochondrial p4502 systems
|