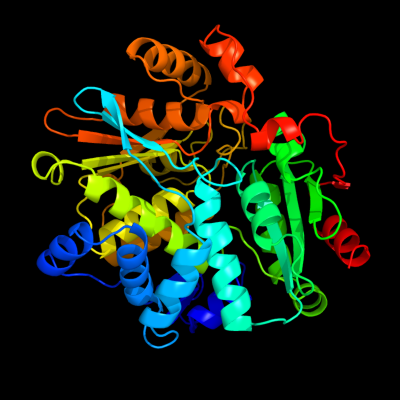
| 1 | d1vb3a1
|
|
 |
100.0 |
100 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
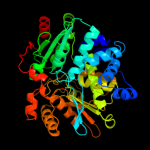
| 2 | d1kl7a_
|
|
 |
100.0 |
35 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
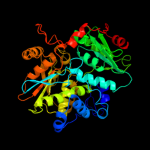
| 3 | c3v7nA_
|
|
 |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase (thrc) from from burkholderia2 thailandensis
|
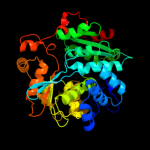
| 4 | d1e5xa_
|
|
 |
100.0 |
23 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 5 | d1v8za1
|
|
 |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 6 | c2d1fA_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: structure of mycobacterium tuberculosis threonine synthase
|
| 7 | d1v7ca_
|
|
 |
100.0 |
24 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 8 | c2zsjB_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
|
| 9 | d1qopb_
|
|
 |
100.0 |
15 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 10 | d1pwha_
|
|
 |
100.0 |
14 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 11 | c1tdjA_
|
|
 |
100.0 |
17 |
PDB header:allostery
Chain: A: PDB Molecule:biosynthetic threonine deaminase;
PDBTitle: threonine deaminase (biosynthetic) from e. coli
|
| 12 | c2rkbE_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: E: PDB Molecule:serine dehydratase-like;
PDBTitle: serine dehydratase like-1 from human cancer cells
|
| 13 | c1x1qA_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
|
| 14 | c2gn0A_
|
|
 |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:threonine dehydratase catabolic;
PDBTitle: crystal structure of dimeric biodegradative threonine deaminase (tdcb)2 from salmonella typhimurium at 1.7 a resolution (triclinic form with3 one complete subunit built in alternate conformation)
|
| 15 | c3r0zA_
|
|
 |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:d-serine dehydratase;
PDBTitle: crystal structure of apo d-serine deaminase from salmonella2 typhimurium
|
| 16 | d1v71a1
|
|
 |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 17 | c3l6cA_
|
|
 |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:serine racemase;
PDBTitle: x-ray crystal structure of rat serine racemase in complex with2 malonate a potent inhibitor
|
| 18 | d1p5ja_
|
|
 |
100.0 |
14 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 19 | c1p5jA_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:l-serine dehydratase;
PDBTitle: crystal structure analysis of human serine dehydratase
|
| 20 | d1tdja1
|
|
 |
100.0 |
19 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 21 | d1y7la1 |
|
not modelled |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 22 | d1wkva1 |
|
not modelled |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 23 | d1ve5a1 |
|
not modelled |
100.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 24 | c3iauA_ |
|
not modelled |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:threonine deaminase;
PDBTitle: the structure of the processed form of threonine deaminase isoform 22 from solanum lycopersicum
|
| 25 | d1ve1a1 |
|
not modelled |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 26 | d1jbqa_ |
|
not modelled |
100.0 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 27 | c1jbqD_ |
|
not modelled |
100.0 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:cystathionine beta-synthase;
PDBTitle: structure of human cystathionine beta-synthase: a unique pyridoxal 5'-2 phosphate dependent hemeprotein
|
| 28 | c3pc3A_ |
|
not modelled |
100.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:cg1753, isoform a;
PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
|
| 29 | d2bhsa1 |
|
not modelled |
100.0 |
19 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 30 | d1z7wa1 |
|
not modelled |
100.0 |
15 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 31 | d1o58a_ |
|
not modelled |
100.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 32 | c3dwgA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase b;
PDBTitle: crystal structure of a sulfur carrier protein complex found in the2 cysteine biosynthetic pathway of mycobacterium tuberculosis
|
| 33 | d1fcja_ |
|
not modelled |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 34 | c2pqmA_ |
|
not modelled |
100.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of cysteine synthase (oass) from entamoeba2 histolytica at 1.86 a resolution
|
| 35 | c2q3bA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase a;
PDBTitle: 1.8 a resolution crystal structure of o-acetylserine sulfhydrylase2 (oass) holoenzyme from mycobacterium tuberculosis
|
| 36 | c2o2jA_ |
|
not modelled |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: mycobacterium tuberculosis tryptophan synthase beta chain2 dimer (apoform)
|
| 37 | c2eguA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:cysteine synthase;
PDBTitle: crystal structure of o-acetylserine sulfhydrase from geobacillus2 kaustophilus hta426
|
| 38 | d1f2da_ |
|
not modelled |
100.0 |
17 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 39 | d1tyza_ |
|
not modelled |
100.0 |
13 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 40 | d1j0aa_ |
|
not modelled |
100.0 |
13 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 41 | c2qx7A_ |
|
not modelled |
80.8 |
18 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
| 42 | d1vp8a_ |
|
not modelled |
80.8 |
17 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 43 | d1xgka_ |
|
not modelled |
79.4 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 44 | d1qsga_ |
|
not modelled |
77.8 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 45 | c3s8mA_ |
|
not modelled |
72.4 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acp reductase;
PDBTitle: the crystal structure of fabv
|
| 46 | d1t57a_ |
|
not modelled |
69.0 |
16 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 47 | c2uv8C_ |
|
not modelled |
68.8 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha (fas2);
PDBTitle: crystal structure of yeast fatty acid synthase with stalled2 acyl carrier protein at 3.1 angstrom resolution
|
| 48 | d1o8ca2 |
|
not modelled |
64.9 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 49 | c3imfA_ |
|
not modelled |
63.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: 1.99 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor'
|
| 50 | d1ecfa1 |
|
not modelled |
63.2 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 51 | d1gph11 |
|
not modelled |
61.0 |
18 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 52 | c3iccA_ |
|
not modelled |
59.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 3-oxoacyl-(acyl carrier protein) reductase;
PDBTitle: crystal structure of a putative 3-oxoacyl-(acyl carrier protein)2 reductase from bacillus anthracis at 1.87 a resolution
|
| 53 | c2q2qG_ |
|
not modelled |
58.5 |
10 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:beta-d-hydroxybutyrate dehydrogenase;
PDBTitle: structure of d-3-hydroxybutyrate dehydrogenase from pseudomonas putida
|
| 54 | c1m6yA_ |
|
not modelled |
58.2 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosyl-methyltransferase mraw;
PDBTitle: crystal structure analysis of tm0872, a putative sam-2 dependent methyltransferase, complexed with sah
|
| 55 | c3ek2D_ |
|
not modelled |
57.8 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of eonyl-(acyl carrier protein) reductase2 from burkholderia pseudomallei 1719b
|
| 56 | c2vkzC_ |
|
not modelled |
57.7 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 57 | c2rgwD_ |
|
not modelled |
55.8 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: catalytic subunit of m. jannaschii aspartate2 transcarbamoylase
|
| 58 | d1ae1a_ |
|
not modelled |
54.7 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 59 | d1q59a_ |
|
not modelled |
52.9 |
11 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
| 60 | c2p2gD_ |
|
not modelled |
50.8 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from mycobacterium2 tuberculosis (rv1656): orthorhombic form
|
| 61 | c3qvoA_ |
|
not modelled |
47.0 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nmra family protein;
PDBTitle: structure of a rossmann-fold nad(p)-binding family protein from2 shigella flexneri.
|
| 62 | c3ezlA_ |
|
not modelled |
46.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:acetoacetyl-coa reductase;
PDBTitle: crystal structure of acetyacetyl-coa reductase from2 burkholderia pseudomallei 1710b
|
| 63 | c1stzB_ |
|
not modelled |
45.4 |
28 |
PDB header:transcription
Chain: B: PDB Molecule:heat-inducible transcription repressor hrca homolog;
PDBTitle: crystal structure of a hypothetical protein at 2.2 a resolution
|
| 64 | c1gph1_ |
|
not modelled |
45.4 |
18 |
PDB header:transferase(glutamine amidotransferase)
Chain: 1: PDB Molecule:glutamine phosphoribosyl-pyrophosphate amidotransferase;
PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
|
| 65 | d2ax3a2 |
|
not modelled |
43.5 |
12 |
Fold:YjeF N-terminal domain-like
Superfamily:YjeF N-terminal domain-like
Family:YjeF N-terminal domain-like |
| 66 | c2qkmF_ |
|
not modelled |
43.4 |
23 |
PDB header:hydrolase
Chain: F: PDB Molecule:spac19a8.12 protein;
PDBTitle: the crystal structure of fission yeast mrna decapping enzyme dcp1-dcp22 complex
|
| 67 | d2qklb1 |
|
not modelled |
43.3 |
23 |
Fold:Dcp2 domain-like
Superfamily:Dcp2 domain-like
Family:Dcp2 box A domain |
| 68 | c3kb8A_ |
|
not modelled |
43.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine phosphoribosyltransferase;
PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
|
| 69 | c3toxG_ |
|
not modelled |
43.2 |
17 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 70 | c3qp9C_ |
|
not modelled |
43.1 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:type i polyketide synthase pikaii;
PDBTitle: the structure of a c2-type ketoreductase from a modular polyketide2 synthase
|
| 71 | d1pq1a_ |
|
not modelled |
41.9 |
14 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
| 72 | c3d3jA_ |
|
not modelled |
41.5 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of human edc3p
|
| 73 | c3grkE_ |
|
not modelled |
41.2 |
13 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
|
| 74 | c2vzkD_ |
|
not modelled |
40.9 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:glutamate n-acetyltransferase 2 beta chain;
PDBTitle: structure of the acyl-enzyme complex of an n-terminal2 nucleophile (ntn) hydrolase, oat2
|
| 75 | d1qyda_ |
|
not modelled |
39.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 76 | c3ijrF_ |
|
not modelled |
37.8 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: 2.05 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor' in complex3 with nad+
|
| 77 | c2vofA_ |
|
not modelled |
37.6 |
25 |
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2-related protein a1;
PDBTitle: structure of mouse a1 bound to the puma bh3-domain
|
| 78 | c3i5mA_ |
|
not modelled |
37.5 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative leucoanthocyanidin reductase 1;
PDBTitle: structure of the apo form of leucoanthocyanidin reductase from vitis2 vinifera
|
| 79 | d1yfza1 |
|
not modelled |
36.9 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 80 | c1yfzA_ |
|
not modelled |
36.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
|
| 81 | d1a1va1 |
|
not modelled |
36.7 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RNA helicase |
| 82 | c3e59A_ |
|
not modelled |
36.5 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:pyoverdine biosynthesis protein pvca;
PDBTitle: crystal structure of the pvca (pa2254) protein from pseudomonas2 aeruginosa
|
| 83 | d1fw8a_ |
|
not modelled |
36.0 |
19 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 84 | c3gdfA_ |
|
not modelled |
35.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable nadp-dependent mannitol dehydrogenase;
PDBTitle: crystal structure of the nadp-dependent mannitol dehydrogenase from2 cladosporium herbarum.
|
| 85 | c1vraB_ |
|
not modelled |
35.1 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:arginine biosynthesis bifunctional protein argj;
PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
|
| 86 | c3v8bC_ |
|
not modelled |
34.8 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier
PDBTitle: crystal structure of a 3-ketoacyl-acp reductase from sinorhizobium2 meliloti 1021
|
| 87 | c3ppiA_ |
|
not modelled |
34.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyacyl-coa dehydrogenase type-2;
PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase type-2 from2 mycobacterium avium
|
| 88 | c1oi7A_ |
|
not modelled |
34.5 |
12 |
PDB header:synthetase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: the crystal structure of succinyl-coa synthetase alpha2 subunit from thermus thermophilus
|
| 89 | c3oidA_ |
|
not modelled |
34.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadph];
PDBTitle: crystal structure of enoyl-acp reductases iii (fabl) from b. subtilis2 (complex with nadp and tcl)
|
| 90 | d1p9oa_ |
|
not modelled |
34.3 |
35 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 91 | d2rhca1 |
|
not modelled |
33.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 92 | c2gn9B_ |
|
not modelled |
33.3 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:udp-glcnac c6 dehydratase;
PDBTitle: crystal structure of udp-glcnac inverting 4,6-dehydratase in complex2 with nadp and udp-glc
|
| 93 | c3it4B_ |
|
not modelled |
33.3 |
32 |
PDB header:transferase
Chain: B: PDB Molecule:arginine biosynthesis bifunctional protein argj
PDBTitle: the crystal structure of ornithine acetyltransferase from2 mycobacterium tuberculosis (rv1653) at 1.7 a
|
| 94 | d1trba1 |
|
not modelled |
32.8 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 95 | c3cxtA_ |
|
not modelled |
32.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase with different specificities;
PDBTitle: quaternary complex structure of gluconate 5-dehydrogenase from2 streptococcus suis type 2
|
| 96 | c2zatC_ |
|
not modelled |
32.5 |
12 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase/reductase sdr family member 4;
PDBTitle: crystal structure of a mammalian reductase
|
| 97 | d1uana_ |
|
not modelled |
32.3 |
23 |
Fold:LmbE-like
Superfamily:LmbE-like
Family:LmbE-like |
| 98 | c3bmrA_ |
|
not modelled |
32.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pteridine reductase;
PDBTitle: structure of pteridine reductase 1 (ptr1) from trypanosoma2 brucei in ternary complex with cofactor (nadp+) and3 inhibitor (compound ax6)
|
| 99 | d1f16a_ |
|
not modelled |
32.0 |
15 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
| 100 | c2gk4A_ |
|
not modelled |
31.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: the crystal structure of the dna/pantothenate metabolism flavoprotein2 from streptococcus pneumoniae
|
| 101 | c2yv6A_ |
|
not modelled |
31.2 |
17 |
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2 homologous antagonist/killer;
PDBTitle: crystal structure of human bcl-2 family protein bak
|
| 102 | d2gnoa2 |
|
not modelled |
30.8 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 103 | d2gdza1 |
|
not modelled |
30.2 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 104 | d1bxla_ |
|
not modelled |
30.0 |
15 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
| 105 | d1xdpa3 |
|
not modelled |
29.8 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 106 | c3ioyB_ |
|
not modelled |
29.4 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: structure of putative short-chain dehydrogenase (saro_0793)2 from novosphingobium aromaticivorans
|
| 107 | c3dfmA_ |
|
not modelled |
29.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:teicoplanin pseudoaglycone deacetylase orf2;
PDBTitle: the crystal structure of the zinc inhibited form of2 teicoplanin deacetylase orf2
|
| 108 | d1hgxa_ |
|
not modelled |
29.0 |
18 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 109 | c1n7gB_ |
|
not modelled |
28.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:gdp-d-mannose-4,6-dehydratase;
PDBTitle: crystal structure of the gdp-mannose 4,6-dehydratase2 ternary complex with nadph and gdp-rhamnose.
|
| 110 | d1hdia_ |
|
not modelled |
28.9 |
21 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 111 | c1w4zA_ |
|
not modelled |
28.8 |
14 |
PDB header:antibiotic biosynthesis
Chain: A: PDB Molecule:ketoacyl reductase;
PDBTitle: structure of actinorhodin polyketide (actiii) reductase
|
| 112 | c2qioA_ |
|
not modelled |
28.6 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: x-ray structure of enoyl-acyl carrier protein reductase from bacillus2 anthracis with triclosan
|
| 113 | d1vz6a_ |
|
not modelled |
28.2 |
20 |
Fold:DmpA/ArgJ-like
Superfamily:DmpA/ArgJ-like
Family:ArgJ-like |
| 114 | c3c7cB_ |
|
not modelled |
27.9 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
|
| 115 | c3dfiA_ |
|
not modelled |
27.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:pseudoaglycone deacetylase dbv21;
PDBTitle: the crystal structure of antimicrobial reagent a40926 pseudoaglycone2 deacetylase dbv21
|
| 116 | c3lxmC_ |
|
not modelled |
27.6 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: 2.00 angstrom resolution crystal structure of a catalytic2 subunit of an aspartate carbamoyltransferase (pyrb) from3 yersinia pestis co92
|
| 117 | c3r3sD_ |
|
not modelled |
27.6 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase;
PDBTitle: structure of the ygha oxidoreductase from salmonella enterica
|
| 118 | d1mxha_ |
|
not modelled |
27.1 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 119 | d1ysga1 |
|
not modelled |
26.9 |
14 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
| 120 | d1k3ka_ |
|
not modelled |
26.8 |
24 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |