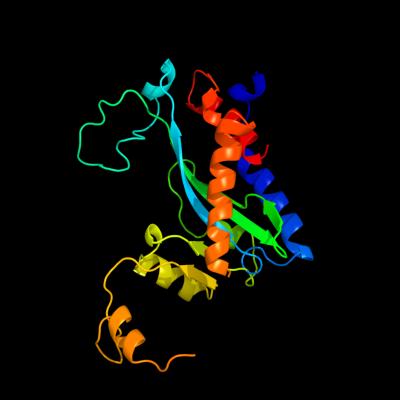
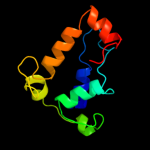
1 d1musa_
98.5
10
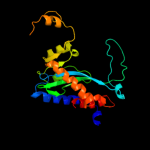
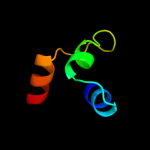
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)2 d1b7ea_
98.1
13
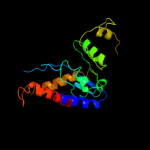
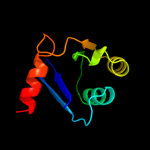
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)3 d1cxqa_
91.1
22
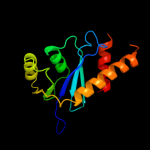
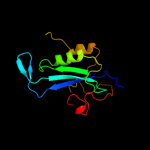
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain4 d1asua_
85.6
17
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain5 d1c0ma2
67.9
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain6 c1bg1A_
66.6
14
PDB header: transcription/dnaChain: A: PDB Molecule: protein (transcription factor stat3b);PDBTitle: transcription factor stat3b/dna complex
7 c3nf9A_
64.5
13
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: integrase;PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
8 c1yvlB_
62.4
14
PDB header: signaling proteinChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: structure of unphosphorylated stat1
9 c3hefB_
51.6
15
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
10 c1c0mA_
47.2
19
PDB header: transferaseChain: A: PDB Molecule: protein (integrase);PDBTitle: crystal structure of rsv two-domain integrase
11 d1bcoa2
46.2
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: mu transposase, core domain12 d1hyva_
40.9
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain13 c1bf5A_
36.9
17
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
14 c3f9kV_
36.4
13
PDB header: viral protein, recombinationChain: V: PDB Molecule: integrase;PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
15 d1exqa_
31.9
16
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain16 c1bcoA_
30.9
16
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
17 d1k78a1
30.8
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain18 c3nzqB_
29.0
17
PDB header: lyaseChain: B: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
19 d6paxa1
24.9
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain20 d1a9xa1
21.6
12
Fold: Carbamoyl phosphate synthetase, large subunit connection domainSuperfamily: Carbamoyl phosphate synthetase, large subunit connection domainFamily: Carbamoyl phosphate synthetase, large subunit connection domain21 c3n2oA_
not modelled
20.4
32
PDB header: lyaseChain: A: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
22 c1k6yB_
not modelled
19.7
15
PDB header: transferaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
23 d1c6va_
not modelled
18.7
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain24 c3iwfA_
not modelled
14.1
4
PDB header: transcription regulatorChain: A: PDB Molecule: transcription regulator rpir family;PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
25 c2jg6A_
not modelled
11.6
9
PDB header: hydrolaseChain: A: PDB Molecule: dna-3-methyladenine glycosidase;PDBTitle: crystal structure of a 3-methyladenine dna glycosylase i2 from staphylococcus aureus
26 d1slma1
not modelled
10.2
27
Fold: PGBD-likeSuperfamily: PGBD-likeFamily: MMP N-terminal domain27 c3nzpA_
not modelled
10.2
25
PDB header: lyaseChain: A: PDB Molecule: arginine decarboxylase;PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
28 c2vkpA_
not modelled
9.7
12
PDB header: protein-bindingChain: A: PDB Molecule: btb/poz domain-containing protein 6;PDBTitle: crystal structure of btb domain from btbd6
29 c3cwgA_
not modelled
9.5
13
PDB header: transcriptionChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: unphosphorylated mouse stat3 core fragment
30 c1y1uA_
not modelled
9.1
16
PDB header: signaling proteinChain: A: PDB Molecule: signal transducer and activator of transcription 5a;PDBTitle: structure of unphosphorylated stat5a
31 c3eusB_
not modelled
8.9
16
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
32 c2o3fC_
not modelled
8.0
12
PDB header: transcriptionChain: C: PDB Molecule: putative hth-type transcriptional regulator ybbh;PDBTitle: structural genomics, the crystal structure of the n-2 terminal domain of the putative transcriptional regulator3 ybbh from bacillus subtilis subsp. subtilis str. 168.
33 d2o3fa1
not modelled
8.0
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: RpiR-like34 d1nkua_
not modelled
8.0
10
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: 3-Methyladenine DNA glycosylase I (Tag)35 c1ex4A_
not modelled
7.7
17
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: hiv-1 integrase catalytic core and c-terminal domain
36 d1a6qa1
not modelled
6.9
27
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain37 c2kvcA_
not modelled
5.8
20
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
38 d1v77a_
not modelled
5.8
7
Fold: 7-stranded beta/alpha barrelSuperfamily: PHP domain-likeFamily: RNase P subunit p3039 d1sbza_
not modelled
5.5
19
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD40 c3hpgC_
not modelled
5.3
13
PDB header: transferaseChain: C: PDB Molecule: integrase;PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
41 c3g7pA_
not modelled
5.2
15
PDB header: unknown functionChain: A: PDB Molecule: nitrogen fixation protein;PDBTitle: crystal structure of a nifx-associated protein of unknown function2 (afe_1514) from acidithiobacillus ferrooxidans atcc at 2.00 a3 resolution
42 c2lm4A_
not modelled
5.1
14
PDB header: protein bindingChain: A: PDB Molecule: succinate dehydrogenase assembly factor 2, mitochondrial;PDBTitle: solution nmr structure of mitochondrial succinate dehydrogenase2 assembly factor 2 from saccharomyces cerevisiae, northeast structural3 genomics consortium target yt682a