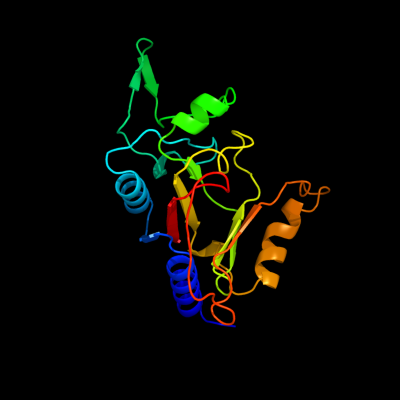
1 d2g40a1
100.0
20
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: YkgG-like2 c2g40A_
100.0
20
PDB header: unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a duf162 family protein (dr_1909) from2 deinococcus radiodurans at 1.70 a resolution
3 d2g82a1
37.6
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain4 d1a5ta2
37.5
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain5 d2pkqo1
33.9
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain6 d1d2na_
33.1
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain7 c1a6cA_
30.7
26
PDB header: virusChain: A: PDB Molecule: tobacco ringspot virus capsid protein;PDBTitle: structure of tobacco ringspot virus
8 d1i32a1
29.4
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain9 d1hdgo1
29.2
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain10 c2y7uM_
28.4
17
PDB header: virusChain: M: PDB Molecule: coat protein;PDBTitle: x-ray structure of the grapevine fanleaf virus
11 c2qz4A_
27.9
13
PDB header: hydrolaseChain: A: PDB Molecule: paraplegin;PDBTitle: human paraplegin, aaa domain in complex with adp
12 d1vc2a1
24.7
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain13 d1obfo1
24.1
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain14 d3gpdg1
23.6
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain15 d3cmco1
21.7
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain16 c1nsfA_
20.7
24
PDB header: protein transportChain: A: PDB Molecule: n-ethylmaleimide sensitive factor;PDBTitle: d2 hexamerization domain of n-ethylmaleimide sensitive factor (nsf)
17 d1u8fo1
20.7
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain18 d1gado1
20.6
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain19 d1nbwa3
18.2
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ATPase domain of dehydratase reactivase alpha subunit20 c3co5B_
17.9
8
PDB header: transcription regulatorChain: B: PDB Molecule: putative two-component system transcriptional responsePDBTitle: crystal structure of sigma-54 interaction domain of putative2 transcriptional response regulator from neisseria gonorrhoeae
21 c2ce7B_
not modelled
17.8
22
PDB header: cell division proteinChain: B: PDB Molecule: cell division protein ftsh;PDBTitle: edta treated
22 d1j0xo1
not modelled
17.4
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain23 d1rm4a1
not modelled
17.1
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain24 c2z1dA_
not modelled
16.5
36
PDB header: metal binding proteinChain: A: PDB Molecule: hydrogenase expression/formation protein hypd;PDBTitle: crystal structure of [nife] hydrogenase maturation protein, hypd from2 thermococcus kodakaraensis
25 c1fnnB_
not modelled
16.4
14
PDB header: cell cycleChain: B: PDB Molecule: cell division control protein 6;PDBTitle: crystal structure of cdc6p from pyrobaculum aerophilum
26 d1q5za_
not modelled
15.8
27
Fold: Invasion protein A (SipA) , C-terminal actin binding domainSuperfamily: Invasion protein A (SipA) , C-terminal actin binding domainFamily: Invasion protein A (SipA) , C-terminal actin binding domain27 c1q5zA_
not modelled
15.8
27
PDB header: cell invasionChain: A: PDB Molecule: sipa;PDBTitle: crystal structure of the c-terminal actin binding domain of2 salmonella invasion protein a (sipa)
28 d1k3ta1
not modelled
15.7
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain29 c3sggA_
not modelled
15.1
28
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical hydrolase;PDBTitle: crystal structure of a hypothetical hydrolase (bt_2193) from2 bacteroides thetaiotaomicron vpi-5482 at 1.25 a resolution
30 c2dhrC_
not modelled
14.4
17
PDB header: hydrolaseChain: C: PDB Molecule: ftsh;PDBTitle: whole cytosolic region of atp-dependent metalloprotease2 ftsh (g399l)
31 c2c9oC_
not modelled
13.4
14
PDB header: hydrolaseChain: C: PDB Molecule: ruvb-like 1;PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
32 c3l7oB_
not modelled
13.3
13
PDB header: isomeraseChain: B: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
33 d1in4a2
not modelled
12.8
8
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain34 c3b9pA_
not modelled
12.7
14
PDB header: hydrolaseChain: A: PDB Molecule: cg5977-pa, isoform a;PDBTitle: spastin
35 c3u7jA_
not modelled
12.5
16
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
36 c1xwiA_
not modelled
12.5
14
PDB header: protein transportChain: A: PDB Molecule: skd1 protein;PDBTitle: crystal structure of vps4b
37 d1sxje2
not modelled
12.3
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain38 d2joya1
not modelled
12.2
14
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e39 d1v5wa_
not modelled
12.2
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)40 c3u5cG_
not modelled
12.1
22
PDB header: ribosomeChain: G: PDB Molecule: 40s ribosomal protein s6-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
41 d2b4ro1
not modelled
11.5
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain42 c1jr3E_
not modelled
11.2
29
PDB header: transferaseChain: E: PDB Molecule: dna polymerase iii, delta' subunit;PDBTitle: crystal structure of the processivity clamp loader gamma2 complex of e. coli dna polymerase iii
43 d1njfa_
not modelled
11.1
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain44 d1ggaa1
not modelled
11.1
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain45 c1w5sB_
not modelled
11.0
13
PDB header: replicationChain: B: PDB Molecule: origin recognition complex subunit 2 orc2;PDBTitle: structure of the aeropyrum pernix orc2 protein (adp form)
46 d2pl1a1
not modelled
10.8
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related47 c2xznY_
not modelled
10.5
17
PDB header: ribosomeChain: Y: PDB Molecule: rps6e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
48 c1ahuB_
not modelled
10.4
15
PDB header: flavoenzymeChain: B: PDB Molecule: vanillyl-alcohol oxidase;PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
49 c1l6jA_
not modelled
10.4
27
PDB header: hydrolaseChain: A: PDB Molecule: matrix metalloproteinase-9;PDBTitle: crystal structure of human matrix metalloproteinase mmp92 (gelatinase b).
50 c2r65A_
not modelled
9.8
12
PDB header: hydrolaseChain: A: PDB Molecule: cell division protease ftsh homolog;PDBTitle: crystal structure of helicobacter pylori atp dependent protease, ftsh2 adp complex
51 d1t9ka_
not modelled
8.9
13
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like52 c2kyrA_
not modelled
8.8
15
PDB header: transferaseChain: A: PDB Molecule: fructose-like phosphotransferase enzyme iib component 1;PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
53 d2r48a1
not modelled
8.7
8
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like54 c3cnbC_
not modelled
8.7
7
PDB header: dna binding proteinChain: C: PDB Molecule: dna-binding response regulator, merr family;PDBTitle: crystal structure of signal receiver domain of dna binding response2 regulator protein (merr) from colwellia psychrerythraea 34h
55 d2a9sa1
not modelled
8.4
11
Fold: Anticodon-binding domain-likeSuperfamily: CinA-likeFamily: CinA-like56 c2zayA_
not modelled
8.3
14
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator from desulfuromonas2 acetoxidans
57 d1u0sy_
not modelled
8.3
7
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related58 d1r7ra3
not modelled
8.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain59 c3eulB_
not modelled
8.1
20
PDB header: transcriptionChain: B: PDB Molecule: possible nitrate/nitrite response transcriptionalPDBTitle: structure of the signal receiver domain of the putative2 response regulator narl from mycobacterium tuberculosis
60 c3d8bB_
not modelled
8.1
17
PDB header: hydrolaseChain: B: PDB Molecule: fidgetin-like protein 1;PDBTitle: crystal structure of human fidgetin-like protein 1 in complex with adp
61 c2j17A_
not modelled
7.6
20
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase yil113w;PDBTitle: ptyr bound form of sdp-1
62 c3q3qA_
not modelled
7.3
13
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
63 c3s93B_
not modelled
7.3
7
PDB header: transcriptionChain: B: PDB Molecule: tudor domain-containing protein 5;PDBTitle: crystal structure of conserved motif in tdrd5
64 d1biaa2
not modelled
7.3
14
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Biotin repressor (BirA)65 d1qvra3
not modelled
7.2
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain66 d1izca_
not modelled
7.1
11
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase67 c1izcA_
not modelled
7.1
11
PDB header: lyaseChain: A: PDB Molecule: macrophomate synthase intermolecular diels-alderase;PDBTitle: crystal structure analysis of macrophomate synthase
68 d1r6bx3
not modelled
7.1
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain69 d1p2fa2
not modelled
7.1
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related70 d1sp8a1
not modelled
6.9
12
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases71 d1lv7a_
not modelled
6.9
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain72 c1eakA_
not modelled
6.7
18
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: 72 kda type iv collagenase;PDBTitle: catalytic domain of prommp-2 e404q mutant
73 d2ce7a2
not modelled
6.6
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain74 c1nbwA_
not modelled
6.5
20
PDB header: hydrolaseChain: A: PDB Molecule: glycerol dehydratase reactivase alpha subunit;PDBTitle: glycerol dehydratase reactivase
75 c3eihB_
not modelled
6.5
14
PDB header: protein transportChain: B: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: crystal structure of s.cerevisiae vps4 in the presence of atpgammas
76 d1fnna2
not modelled
6.3
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain77 d1zesa1
not modelled
6.2
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related78 d1sqia1
not modelled
6.2
10
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases79 c3ecjC_
not modelled
6.1
38
PDB header: oxidoreductaseChain: C: PDB Molecule: protein (homoprotocatechuate 2,3-dioxygenase);PDBTitle: structure of e323l mutant of homoprotocatechuate 2,3-dioxygenase from2 brevibacterium fuscum at 1.65a resolution
80 c3b8iF_
not modelled
6.1
21
PDB header: lyaseChain: F: PDB Molecule: pa4872 oxaloacetate decarboxylase;PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
81 c3iuyB_
not modelled
6.1
18
PDB header: hydrolaseChain: B: PDB Molecule: probable atp-dependent rna helicase ddx53;PDBTitle: crystal structure of ddx53 dead-box domain
82 d1e94e_
not modelled
5.9
38
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain83 d1stza2
not modelled
5.7
11
Fold: Profilin-likeSuperfamily: GAF domain-likeFamily: HrcA C-terminal domain-like84 d2d0oa3
not modelled
5.6
20
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ATPase domain of dehydratase reactivase alpha subunit85 c4a1dE_
not modelled
5.6
25
PDB header: ribosomeChain: E: PDB Molecule: rpl6;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
86 c3izcG_
not modelled
5.3
36
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein rpl6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
87 d1s8na_
not modelled
5.3
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related88 c2yvkA_
not modelled
5.2
12
PDB header: isomeraseChain: A: PDB Molecule: methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
89 d1g6oa_
not modelled
5.2
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)90 c2f6rA_
not modelled
5.2
14
PDB header: transferaseChain: A: PDB Molecule: bifunctional coenzyme a synthase;PDBTitle: crystal structure of bifunctional coenzyme a synthase (coa synthase):2 (18044849) from mus musculus at 1.70 a resolution
91 d1sp8a2
not modelled
5.1
11
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases