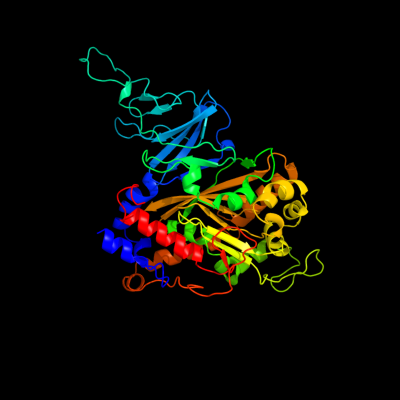
| 1 | c2gbxE_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:biphenyl 2,3-dioxygenase alpha subunit;
PDBTitle: crystal structure of biphenyl 2,3-dioxygenase from sphingomonas2 yanoikuyae b1 bound to biphenyl
|
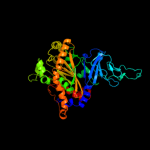
| 2 | c2hmnA_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:naphthalene 1,2-dioxygenase alpha subunit;
PDBTitle: crystal structure of the naphthalene 1,2-dioxygenase f352v2 mutant bound to anthracene.
|
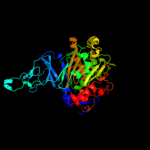
| 3 | c1wqlA_
|
|
 |
100.0 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-sulfur protein large subunit of cumene dioxygenase;
PDBTitle: cumene dioxygenase (cuma1a2) from pseudomonas fluorescens ip01
|
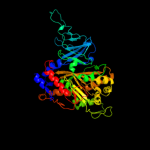
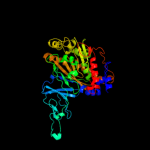
| 4 | c1uljA_
|
|
 |
100.0 |
48 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biphenyl dioxygenase large subunit;
PDBTitle: biphenyl dioxygenase (bpha1a2) in complex with the substrate
|
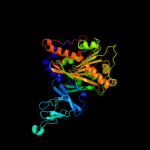
| 5 | c2b1xE_
|
|
 |
100.0 |
39 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:naphthalene dioxygenase large subunit;
PDBTitle: crystal structure of naphthalene 1,2-dioxygenase from rhodococcus sp.
|
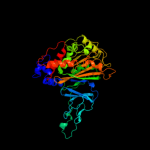
| 6 | c3n0qA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative aromatic-ring hydroxylating dioxygenase;
PDBTitle: crystal structure of a putative aromatic-ring hydroxylating2 dioxygenase (tm1040_3219) from silicibacter sp. tm1040 at 1.80 a3 resolution
|
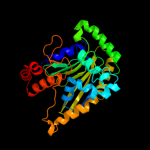
| 7 | d1o7na2
|
|
 |
100.0 |
30 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 8 | d1wqla2
|
|
 |
100.0 |
38 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 9 | d2bmoa2
|
|
 |
100.0 |
30 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 10 | d1ulia2
|
|
 |
100.0 |
44 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 11 | d2b1xa2
|
|
 |
100.0 |
34 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 12 | c3gteB_
|
|
 |
100.0 |
16 |
PDB header:electron transport, oxidoreductase
Chain: B: PDB Molecule:ddmc;
PDBTitle: crystal structure of dicamba monooxygenase with non-heme2 iron
|
| 13 | c3gkqB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:terminal oxygenase component of carbazole 1,9a-
PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 novosphingobium sp. ka1
|
| 14 | c3gcfC_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:terminal oxygenase component of carbazole 1,9a-
PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 nocardioides aromaticivorans ic177
|
| 15 | d1wqla1
|
|
 |
100.0 |
56 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 16 | c1z01D_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:2-oxo-1,2-dihydroquinoline 8-monooxygenase,
PDBTitle: 2-oxoquinoline 8-monooxygenase component: active site2 modulation by rieske-[2fe-2s] center oxidation/reduction
|
| 17 | d2b1xa1
|
|
 |
100.0 |
48 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 18 | d1ulia1
|
|
 |
100.0 |
58 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 19 | d2bmoa1
|
|
 |
100.0 |
47 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 20 | c2zylA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:possible oxidoreductase;
PDBTitle: crystal structure of 3-ketosteroid-9-alpha-hydroxylase2 (ksha) from m. tuberculosis
|
| 21 | c2de7B_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:terminal oxygenase component of carbazole;
PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
|
| 22 | d1o7na1 |
|
not modelled |
100.0 |
46 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 23 | d1z01a1 |
|
not modelled |
100.0 |
21 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 24 | d2de6a1 |
|
not modelled |
100.0 |
25 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 25 | c3d89A_ |
|
not modelled |
99.9 |
16 |
PDB header:electron transport
Chain: A: PDB Molecule:rieske domain-containing protein;
PDBTitle: crystal structure of a soluble rieske ferredoxin from mus musculus
|
| 26 | d1vm9a_ |
|
not modelled |
99.8 |
21 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 27 | c2de7E_ |
|
not modelled |
99.8 |
21 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ferredoxin component of carbazole;
PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
|
| 28 | d2jzaa1 |
|
not modelled |
99.8 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 29 | c3dqyA_ |
|
not modelled |
99.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:toluene 1,2-dioxygenase system ferredoxin
PDBTitle: crystal structure of toluene 2,3-dioxygenase ferredoxin
|
| 30 | d3c0da1 |
|
not modelled |
99.8 |
12 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 31 | c3gceA_ |
|
not modelled |
99.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin component of carbazole 1,9a-
PDBTitle: ferredoxin of carbazole 1,9a-dioxygenase from nocardioides2 aromaticivorans ic177
|
| 32 | d1fqta_ |
|
not modelled |
99.8 |
16 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 33 | c2qpzA_ |
|
not modelled |
99.8 |
22 |
PDB header:metal binding protein
Chain: A: PDB Molecule:naphthalene 1,2-dioxygenase system ferredoxin
PDBTitle: naphthalene 1,2-dioxygenase rieske ferredoxin
|
| 34 | d2jo6a1 |
|
not modelled |
99.8 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 35 | c2i7fB_ |
|
not modelled |
99.7 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin component of dioxygenase;
PDBTitle: sphingomonas yanoikuyae b1 ferredoxin
|
| 36 | d1q90c_ |
|
not modelled |
99.6 |
23 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 37 | d1rfsa_ |
|
not modelled |
99.6 |
24 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 38 | d3cx5e1 |
|
not modelled |
99.4 |
28 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 39 | d2e74d1 |
|
not modelled |
99.4 |
22 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 40 | d1g8kb_ |
|
not modelled |
99.4 |
18 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 41 | d1riea_ |
|
not modelled |
99.4 |
23 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 42 | c2nvgA_ |
|
not modelled |
99.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: soluble domain of rieske iron sulfur protein.
|
| 43 | c2e76D_ |
|
not modelled |
99.0 |
23 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
| 44 | c2fynO_ |
|
not modelled |
99.0 |
21 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
| 45 | d1nyka_ |
|
not modelled |
98.9 |
24 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 46 | c2fyuE_ |
|
not modelled |
98.8 |
22 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
| 47 | c1p84E_ |
|
not modelled |
98.3 |
25 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
| 48 | d1z01a2 |
|
not modelled |
97.7 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 49 | d1jm1a_ |
|
not modelled |
97.3 |
25 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
| 50 | d2de6a2 |
|
not modelled |
96.3 |
17 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Ring hydroxylating alpha subunit catalytic domain |
| 51 | d1uhva1 |
|
not modelled |
31.4 |
28 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:Composite domain of glycosyl hydrolase families 5, 30, 39 and 51 |
| 52 | c3fcgB_ |
|
not modelled |
29.7 |
27 |
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:f1 capsule-anchoring protein;
PDBTitle: crystal structure analysis of the middle domain of the2 caf1a usher
|
| 53 | c3kv0A_ |
|
not modelled |
28.3 |
43 |
PDB header:transport protein
Chain: A: PDB Molecule:het-c2;
PDBTitle: crystal structure of het-c2: a fungal glycolipid transfer protein2 (gltp)
|
| 54 | d1mkna_ |
|
not modelled |
24.1 |
67 |
Fold:Midkine
Superfamily:Midkine
Family:Midkine, a heparin-binding growth factor, N-terminal domain |
| 55 | d1wmxa_ |
|
not modelled |
23.9 |
22 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Family 30 carbohydrate binding module, CBM30 (PKD repeat) |
| 56 | d2fu5a1 |
|
not modelled |
23.8 |
25 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:RabGEF Mss4 |
| 57 | d1pwka_ |
|
not modelled |
23.7 |
13 |
Fold:DLC
Superfamily:DLC
Family:DLC |
| 58 | c1ddzA_ |
|
not modelled |
23.2 |
42 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: x-ray structure of a beta-carbonic anhydrase from the red2 alga, porphyridium purpureum r-1
|
| 59 | c1lshB_ |
|
not modelled |
22.3 |
19 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:lipovitellin (lv-2);
PDBTitle: lipid-protein interactions in lipovitellin
|
| 60 | d1lshb_ |
|
not modelled |
22.3 |
19 |
Fold:Lipovitellin-phosvitin complex; beta-sheet shell regions
Superfamily:Lipovitellin-phosvitin complex; beta-sheet shell regions
Family:Lipovitellin-phosvitin complex; beta-sheet shell regions |
| 61 | d1w91a1 |
|
not modelled |
22.2 |
50 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:Composite domain of glycosyl hydrolase families 5, 30, 39 and 51 |
| 62 | d1hxra_ |
|
not modelled |
21.5 |
25 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:RabGEF Mss4 |
| 63 | d3brda2 |
|
not modelled |
18.6 |
35 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:p53-like transcription factors
Family:DNA-binding protein LAG-1 (CSL) |
| 64 | d1ddza2 |
|
not modelled |
18.6 |
42 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 65 | c2x3mA_ |
|
not modelled |
18.4 |
33 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein orf239;
PDBTitle: crystal structure of hypothetical protein orf239 from pyrobaculum2 spherical virus
|
| 66 | c2wb6A_ |
|
not modelled |
18.0 |
16 |
PDB header:viral protein
Chain: A: PDB Molecule:afv1-102;
PDBTitle: crystal structure of afv1-102, a protein from the acidianus2 filamentous virus 1
|
| 67 | d3orca_ |
|
not modelled |
16.7 |
24 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Phage repressors |
| 68 | c2c3yA_ |
|
not modelled |
16.3 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate-ferredoxin oxidoreductase;
PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
|
| 69 | c3lasA_ |
|
not modelled |
15.5 |
29 |
PDB header:lyase
Chain: A: PDB Molecule:putative carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
|
| 70 | d1nr3a_ |
|
not modelled |
15.3 |
19 |
Fold:DNA-binding protein Tfx
Superfamily:DNA-binding protein Tfx
Family:DNA-binding protein Tfx |
| 71 | c1ylkA_ |
|
not modelled |
14.4 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein rv1284/mt1322;
PDBTitle: crystal structure of rv1284 from mycobacterium tuberculosis in complex2 with thiocyanate
|
| 72 | d1swxa_ |
|
not modelled |
14.1 |
80 |
Fold:Glycolipid transfer protein, GLTP
Superfamily:Glycolipid transfer protein, GLTP
Family:Glycolipid transfer protein, GLTP |
| 73 | c2i3fA_ |
|
not modelled |
13.5 |
40 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:glycolipid transfer-like protein;
PDBTitle: crystal structure of a glycolipid transfer-like protein2 from galdieria sulphuraria
|
| 74 | d1cmia_ |
|
not modelled |
13.5 |
13 |
Fold:DLC
Superfamily:DLC
Family:DLC |
| 75 | d1e1oa1 |
|
not modelled |
13.1 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 76 | c2lm4A_ |
|
not modelled |
13.0 |
38 |
PDB header:protein binding
Chain: A: PDB Molecule:succinate dehydrogenase assembly factor 2, mitochondrial;
PDBTitle: solution nmr structure of mitochondrial succinate dehydrogenase2 assembly factor 2 from saccharomyces cerevisiae, northeast structural3 genomics consortium target yt682a
|
| 77 | d2c42a2 |
|
not modelled |
13.0 |
27 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:PFOR PP module |
| 78 | c2jz8A_ |
|
not modelled |
12.6 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bh09830;
PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
|
| 79 | c2vpyB_ |
|
not modelled |
12.4 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nrfc protein;
PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
|
| 80 | c3brgC_ |
|
not modelled |
12.2 |
35 |
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:recombining binding protein suppressor of
PDBTitle: csl (rbp-jk) bound to dna
|
| 81 | d3cdda1 |
|
not modelled |
12.2 |
17 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
| 82 | c3izbP_ |
|
not modelled |
12.1 |
60 |
PDB header:ribosome
Chain: P: PDB Molecule:40s ribosomal protein rps11 (s17p);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 83 | c2a8cE_ |
|
not modelled |
11.9 |
50 |
PDB header:lyase
Chain: E: PDB Molecule:carbonic anhydrase 2;
PDBTitle: haemophilus influenzae beta-carbonic anhydrase
|
| 84 | c3iz6P_ |
|
not modelled |
11.8 |
60 |
PDB header:ribosome
Chain: P: PDB Molecule:40s ribosomal protein s11 (s17p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 85 | d3e2ba1 |
|
not modelled |
11.8 |
13 |
Fold:DLC
Superfamily:DLC
Family:DLC |
| 86 | c2fo1A_ |
|
not modelled |
11.6 |
35 |
PDB header:gene regulation/signalling protein/dna
Chain: A: PDB Molecule:lin-12 and glp-1 phenotype protein 1, isoform b;
PDBTitle: crystal structure of the csl-notch-mastermind ternary2 complex bound to dna
|
| 87 | d1bbua1 |
|
not modelled |
11.4 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Anticodon-binding domain |
| 88 | d1kt0a2 |
|
not modelled |
11.2 |
26 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 89 | d1ddza1 |
|
not modelled |
10.9 |
43 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 90 | c2yrtA_ |
|
not modelled |
10.8 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:chord containing protein-1;
PDBTitle: solution structure of the chord domain of human chord-2 containing protein 1
|
| 91 | c2w3nA_ |
|
not modelled |
10.8 |
32 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase 2;
PDBTitle: structure and inhibition of the co2-sensing carbonic2 anhydrase can2 from the pathogenic fungus cryptococcus3 neoformans
|
| 92 | d1nfga1 |
|
not modelled |
10.6 |
22 |
Fold:Composite domain of metallo-dependent hydrolases
Superfamily:Composite domain of metallo-dependent hydrolases
Family:Hydantoinase (dihydropyrimidinase) |
| 93 | d1ubdc4 |
|
not modelled |
10.5 |
44 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 94 | d1vmka_ |
|
not modelled |
10.4 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 95 | c3eyxB_ |
|
not modelled |
10.2 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
|
| 96 | d2abxa_ |
|
not modelled |
10.2 |
42 |
Fold:Snake toxin-like
Superfamily:Snake toxin-like
Family:Snake venom toxins |
| 97 | d1d1la_ |
|
not modelled |
10.2 |
24 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Phage repressors |
| 98 | c2ivfB_ |
|
not modelled |
9.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ethylbenzene dehydrogenase beta-subunit;
PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
|
| 99 | c2xznQ_ |
|
not modelled |
9.6 |
40 |
PDB header:ribosome
Chain: Q: PDB Molecule:ribosomal protein s17 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|