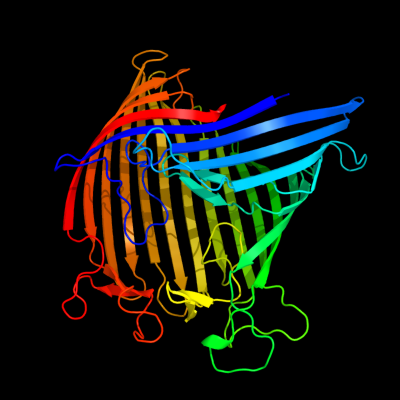
1 d1af6a_
100.0
100
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like2 d2mpra_
100.0
80
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like3 d1a0tp_
100.0
29
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like4 c3qraA_
96.9
11
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
5 c2vdaB_
95.8
100
PDB header: protein transportChain: B: PDB Molecule: maltoporin;PDBTitle: solution structure of the seca-signal peptide complex
6 c3nsgA_
93.7
12
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
7 d2zfga1
93.1
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin8 d1qj8a_
92.7
13
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein9 d1phoa_
91.9
9
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin10 d1osma_
88.8
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin11 c2wjqA_
82.8
14
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
12 c3nb3C_
79.4
10
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
13 d1qjpa_
79.1
10
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein14 d1g90a_
73.7
12
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein15 c2x4mD_
60.4
21
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
16 d1i78a_
59.4
17
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane protease OMPT17 c2f1tB_
59.1
8
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
18 c2k0lA_
57.5
11
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
19 c2jmmA_
38.2
13
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
20 d2vdfa1
37.3
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane adhesin/invasin OpcA21 c3dzmB_
not modelled
35.6
19
PDB header: unknown functionChain: B: PDB Molecule: hypothetical conserved protein;PDBTitle: crystal structure of a major outer membrane protein from thermus2 thermophilus hb27
22 c2iwvD_
not modelled
25.0
14
PDB header: ion channelChain: D: PDB Molecule: outer membrane protein g;PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
23 c2o4vA_
not modelled
25.0
17
PDB header: membrane proteinChain: A: PDB Molecule: porin p;PDBTitle: an arginine ladder in oprp mediates phosphate specific transfer across2 the outer membrane
24 d2fgqx1
not modelled
23.6
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin25 c2l8aA_
not modelled
20.7
14
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: structure of a novel cbm3 lacking the calcium-binding site
26 d2jnaa1
not modelled
18.6
33
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like27 c2bs5A_
not modelled
12.7
35
PDB header: sugar binding proteinChain: A: PDB Molecule: fucose-binding lectin protein;PDBTitle: lectin from ralstonia solanacearum complexed with 2-2 fucosyllactose
28 d1cz5a2
not modelled
9.3
50
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like29 c2wo4A_
not modelled
9.3
16
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase, family 9;PDBTitle: 3b' carbohydrate-binding module from the cel9v glycoside2 hydrolase from clostridium thermocellum, in-house data
30 c3aehB_
not modelled
8.8
9
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
31 c2rfyB_
not modelled
8.1
20
PDB header: hydrolaseChain: B: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: crystal structure of cellobiohydrolase from melanocarpus2 albomyces complexed with cellobiose
32 d2jb0b1
not modelled
8.0
29
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif33 c3sf5D_
not modelled
7.7
9
PDB header: chaperoneChain: D: PDB Molecule: urease accessory protein ureh;PDBTitle: crystal structure of helicobacter pylori urease accessory protein2 uref/h complex
34 d1p4ta_
not modelled
7.4
8
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein35 c2x27X_
not modelled
7.0
12
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
36 d3prna_
not modelled
6.7
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin37 c3lywA_
not modelled
6.3
39
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ybbr family protein;PDBTitle: crystal structure of ybbr family protein dhaf_0833 from2 desulfitobacterium hafniense dcb-2. northeast structural3 genomics consortium target id dhr29b
38 d2q07a3
not modelled
6.2
14
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: AF0587 pre C-terminal domain-like39 d2v3ia1
not modelled
6.0
24
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core40 c3dwoX_
not modelled
5.8
15
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
41 c3qq2C_
not modelled
5.8
11
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
42 c2outA_
not modelled
5.7
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mu-like prophage flumu protein gp35, proteinPDBTitle: solution structure of hi1506, a novel two domain protein2 from haemophilus influenzae
43 c3a2rX_
not modelled
5.7
11
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
44 c3kvnA_
not modelled
5.7
12
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
45 d1g43a_
not modelled
5.6
12
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III46 d1tpna_
not modelled
5.6
57
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module47 d1nbca_
not modelled
5.4
14
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III48 c3kihC_
not modelled
5.3
21
PDB header: sugar binding proteinChain: C: PDB Molecule: 5-bladed -propeller lectin;PDBTitle: the crystal structures of two fragments truncated from 5-bladed -2 propeller lectin, tachylectin-2 (lib2-d2-15)
49 d1ojja_
not modelled
5.1
22
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core50 c2ei5B_
not modelled
5.1
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ttha0061;PDBTitle: crystal structure of hypothetical protein(ttha0061) from thermus2 thermophilus
51 d2outa2
not modelled
5.1
31
Fold: GINS/PriA/YqbF domainSuperfamily: PriA/YqbF domainFamily: YqbF N-terminal domain-like52 d3ovwa_
not modelled
5.1
13
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core