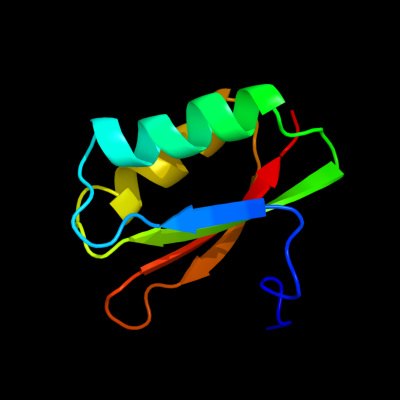
1 d1dcja_
99.9
100
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like2 d1je3a_
99.9
30
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like3 d1pava_
99.8
27
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like4 d1jdqa_
99.8
30
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like5 c3hz7A_
99.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the sira-like protein (dsy4693) from2 desulfitobacterium hafniense, northeast structural genomics3 consortium target dhr2a
6 c1yg0A_
90.5
22
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
7 c1y3kA_
86.0
13
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the fifth domain of2 menkes protein
8 d1p6ta1
82.0
20
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain9 d1p6ta2
79.7
15
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain10 d1kvja_
78.4
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain11 c2gcfA_
74.5
11
PDB header: hydrolaseChain: A: PDB Molecule: cation-transporting atpase pacs;PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
12 d2d9ia1
73.0
12
Fold: IF3-likeSuperfamily: SMR domain-likeFamily: Smr domain13 d1cc8a_
72.9
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain14 c2kt2A_
71.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
15 c3qwuA_
70.3
9
PDB header: ligaseChain: A: PDB Molecule: dna ligase;PDBTitle: putative atp-dependent dna ligase from aquifex aeolicus.
16 d2aw0a_
70.3
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain17 d1q8la_
70.2
10
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain18 c3grzA_
68.3
12
PDB header: transferaseChain: A: PDB Molecule: ribosomal protein l11 methyltransferase;PDBTitle: crystal structure of ribosomal protein l11 methylase from2 lactobacillus delbrueckii subsp. bulgaricus
19 c2vkcA_
64.9
13
PDB header: hydrolaseChain: A: PDB Molecule: nedd4-binding protein 2;PDBTitle: solution structure of the b3bp smr domain
20 d2ggpb1
64.8
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain21 d1s6ua_
not modelled
64.3
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain22 c2l3mA_
not modelled
61.4
21
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
23 c1yjrA_
not modelled
60.0
17
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
24 d2qifa1
not modelled
55.5
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain25 d1mwza_
not modelled
54.2
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain26 c2ldiA_
not modelled
51.7
13
PDB header: hydrolaseChain: A: PDB Molecule: zinc-transporting atpase;PDBTitle: nmr solution structure of ziaan sub mutant
27 c2kyzA_
not modelled
51.3
13
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: nmr structure of heavy metal binding protein tm0320 from thermotoga2 maritima
28 d1cpza_
not modelled
48.4
13
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain29 c3dxsX_
not modelled
48.2
14
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
30 c2aivA_
not modelled
47.3
12
PDB header: transport proteinChain: A: PDB Molecule: fragment of nucleoporin nup116/nsp116;PDBTitle: multiple conformations in the ligand-binding site of the2 yeast nuclear pore targeting domain of nup116p
31 d1osda_
not modelled
46.1
15
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain32 d2nxca1
not modelled
40.8
7
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Ribosomal protein L11 methyltransferase PrmA33 c2ofhX_
not modelled
38.0
11
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
34 c3nf5A_
not modelled
37.9
6
PDB header: protein transportChain: A: PDB Molecule: nucleoporin nup116;PDBTitle: crystal structure of the c-terminal domain of nuclear pore complex2 component nup116 from candida glabrata
35 d1afia_
not modelled
36.9
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain36 d1sb6a_
not modelled
35.2
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain37 c1ko6A_
not modelled
34.9
0
PDB header: transferaseChain: A: PDB Molecule: nuclear pore complex protein nup98;PDBTitle: crystal structure of c-terminal autoproteolytic domain of2 nucleoporin nup98
38 c2q5xA_
not modelled
32.8
0
PDB header: protein transportChain: A: PDB Molecule: nuclear pore complex protein nup98;PDBTitle: crystal structure of the c-terminal domain of hnup98
39 c2kkhA_
not modelled
32.4
13
PDB header: metal transportChain: A: PDB Molecule: putative heavy metal transporter;PDBTitle: structure of the zinc binding domain of the atpase hma4
40 c3cg6A_
not modelled
30.6
23
PDB header: cell cycleChain: A: PDB Molecule: growth arrest and dna-damage-inducible 45 gamma;PDBTitle: crystal structure of gadd45 gamma
41 c3kepA_
not modelled
29.7
6
PDB header: protein transport, rna binding proteinChain: A: PDB Molecule: nucleoporin nup145;PDBTitle: crystal structure of the autoproteolytic domain from the2 nuclear pore complex component nup145 from saccharomyces3 cerevisiae
42 c3qd7X_
not modelled
29.3
21
PDB header: hydrolaseChain: X: PDB Molecule: uncharacterized protein ydal;PDBTitle: crystal structure of ydal, a stand-alone small muts-related protein2 from escherichia coli
43 c2zqeA_
not modelled
27.4
21
PDB header: dna binding proteinChain: A: PDB Molecule: muts2 protein;PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
44 c2k2pA_
not modelled
25.4
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
45 d1dusa_
not modelled
24.9
4
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Hypothetical protein MJ088246 d1fe0a_
not modelled
24.7
11
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain47 c3ntaA_
not modelled
24.4
11
PDB header: oxidoreductaseChain: A: PDB Molecule: fad-dependent pyridine nucleotide-disulphidePDBTitle: structure of the shewanella loihica pv-4 nadh-dependent persulfide2 reductase
48 d1y8ca_
not modelled
23.9
24
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: CAC2371-like49 d1uv7a_
not modelled
22.9
15
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: General secretion pathway protein M, EpsMFamily: General secretion pathway protein M, EpsM50 c1uv7A_
not modelled
22.9
15
PDB header: transportChain: A: PDB Molecule: general secretion pathway protein m;PDBTitle: periplasmic domain of epsm from vibrio cholerae
51 d1wj4a_
not modelled
22.8
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain52 c2kngA_
not modelled
22.7
16
PDB header: dna binding proteinChain: A: PDB Molecule: protein lsr2;PDBTitle: solution structure of c-domain of lsr2
53 d1h8ca_
not modelled
20.5
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain54 c2rmlA_
not modelled
17.2
16
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting p-type atpase copa;PDBTitle: solution structure of the n-terminal soluble domains of2 bacillus subtilis copa
55 d2cr5a1
not modelled
17.1
7
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain56 d1jvaa3
not modelled
17.1
9
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease57 c2rogA_
not modelled
16.2
18
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
58 c3fpnB_
not modelled
16.1
3
PDB header: dna binding proteinChain: B: PDB Molecule: geobacillus stearothermophilus uvrb interactionPDBTitle: crystal structure of uvra-uvrb interaction domains
59 c2ziuA_
not modelled
15.8
8
PDB header: hydrolaseChain: A: PDB Molecule: mus81 protein;PDBTitle: crystal structure of the mus81-eme1 complex
60 d2cc0a1
not modelled
15.1
11
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase61 d1i42a_
not modelled
13.2
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain62 c3qq5A_
not modelled
12.1
17
PDB header: oxidoreductaseChain: A: PDB Molecule: small gtp-binding protein;PDBTitle: crystal structure of the [fefe]-hydrogenase maturation protein hydf
63 c3icrA_
not modelled
11.7
7
PDB header: oxidoreductaseChain: A: PDB Molecule: coenzyme a-disulfide reductase;PDBTitle: crystal structure of oxidized bacillus anthracis coadr-rhd
64 c3dlcA_
not modelled
11.3
6
PDB header: transferaseChain: A: PDB Molecule: putative s-adenosyl-l-methionine-dependentPDBTitle: crystal structure of a putative s-adenosyl-l-methionine-dependent2 methyltransferase (mmp1179) from methanococcus maripaludis at 1.15 a3 resolution
65 d1j23a_
not modelled
11.1
14
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: XPF/Rad1/Mus81 nuclease66 c3ndjA_
not modelled
11.1
24
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: x-ray structure of a c-3'-methyltransferase in complex with s-2 adenosyl-l-homocysteine and sugar product
67 d1l3ia_
not modelled
11.0
14
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Precorrin-6Y methyltransferase (CbiT)68 c2jraB_
not modelled
10.9
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein rpa2121;PDBTitle: a novel domain-swapped solution nmr structure of protein rpa2121 from2 rhodopseudomonas palustris. northeast structural genomics target rpt6
69 c2ga7A_
not modelled
10.9
13
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
70 d2naca2
not modelled
10.8
15
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain71 c3lteH_
not modelled
10.8
14
PDB header: transcriptionChain: H: PDB Molecule: response regulator;PDBTitle: crystal structure of response regulator (signal receiver domain) from2 bermanella marisrubri
72 d2obba1
not modelled
10.3
15
Fold: HAD-likeSuperfamily: HAD-likeFamily: BT0820-like73 c2pwyB_
not modelled
10.0
13
PDB header: transferaseChain: B: PDB Molecule: trna (adenine-n(1)-)-methyltransferase;PDBTitle: crystal structure of a m1a58 trna methyltransferase
74 c2kxjA_
not modelled
9.4
11
PDB header: protein bindingChain: A: PDB Molecule: ubx domain-containing protein 4;PDBTitle: solution structure of ubx domain of human ubxd2 protein
75 c3fryB_
not modelled
9.3
18
PDB header: hydrolaseChain: B: PDB Molecule: probable copper-exporting p-type atpase a;PDBTitle: crystal structure of the copa c-terminal metal binding domain
76 c2zpmA_
not modelled
9.3
13
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain b
77 c3d00A_
not modelled
9.1
22
PDB header: metal binding proteinChain: A: PDB Molecule: tungsten formylmethanofuran dehydrogenase subunit e;PDBTitle: crystal structure of a tungsten formylmethanofuran dehydrogenase2 subunit e (fmde)-like protein (syn_00638) from syntrophus3 aciditrophicus at 1.90 a resolution
78 c2eseA_
not modelled
8.9
22
PDB header: protein/rna complexChain: A: PDB Molecule: vts1p;PDBTitle: structure of the sam domain of vts1p in complex with rna
79 c2b6gA_
not modelled
8.8
22
PDB header: rna binding proteinChain: A: PDB Molecule: vts1p;PDBTitle: rna recognition by the vts1 sam domain
80 d1s3sg_
not modelled
8.5
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain81 c2ropA_
not modelled
8.3
11
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: solution structure of domains 3 and 4 of human atp7b
82 c3evzA_
not modelled
8.1
13
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: crystal strucure of methyltransferase from pyrococcus furiosus
83 d2bgwa2
not modelled
8.0
13
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: XPF/Rad1/Mus81 nuclease84 c3iz6A_
not modelled
7.8
8
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein sa (s2p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
85 d2b2na1
not modelled
7.6
8
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain86 c3hm2G_
not modelled
7.5
7
PDB header: transferaseChain: G: PDB Molecule: precorrin-6y c5,15-methyltransferase;PDBTitle: crystal structure of putative precorrin-6y c5,15-2 methyltransferase targeted domain from corynebacterium3 diphtheriae
87 c3njrB_
not modelled
7.1
10
PDB header: transferaseChain: B: PDB Molecule: precorrin-6y methylase;PDBTitle: crystal structure of c-terminal domain of precorrin-6y c5,15-2 methyltransferase from rhodobacter capsulatus
88 c3c5yD_
not modelled
7.0
10
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
89 d1o54a_
not modelled
6.7
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like90 d1qupa2
not modelled
6.4
16
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain91 c2dzkA_
not modelled
6.4
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ubx domain-containing protein 2;PDBTitle: structure of the ubx domain in mouse ubx domain-containing2 protein 2
92 c3hjhA_
not modelled
6.3
8
PDB header: hydrolaseChain: A: PDB Molecule: transcription-repair-coupling factor;PDBTitle: a rigid n-terminal clamp restrains the motor domains of the bacterial2 transcription-repair coupling factor
93 c3mruB_
not modelled
6.1
16
PDB header: hydrolaseChain: B: PDB Molecule: aminoacyl-histidine dipeptidase;PDBTitle: crystal structure of aminoacylhistidine dipeptidase from vibrio2 alginolyticus
94 c3fpnA_
not modelled
5.9
8
PDB header: dna binding proteinChain: A: PDB Molecule: geobacillus stearothermophilus uvra interactionPDBTitle: crystal structure of uvra-uvrb interaction domains
95 c2aj1A_
not modelled
5.9
14
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
96 d2b25a1
not modelled
5.8
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like97 d1u0sy_
not modelled
5.7
23
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related98 c2rpiA_
not modelled
5.7
6
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease h;PDBTitle: the nmr structure of the submillisecond folding2 intermediate of the thermus thermophilus ribonuclease h
99 d2ciwa1
not modelled
5.6
38
Fold: EF Hand-likeSuperfamily: CloroperoxidaseFamily: Cloroperoxidase