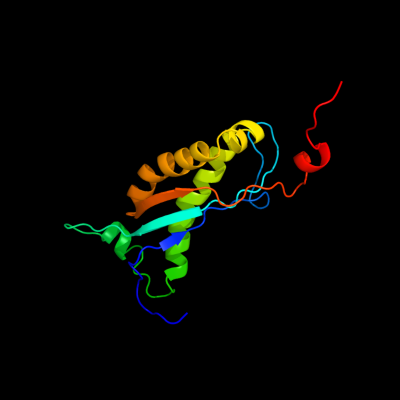
1 d1wina_
99.8
16
Fold: EF-Ts domain-likeSuperfamily: Band 7/SPFH domainFamily: Band 7/SPFH domain2 c3bk6C_
99.8
21
PDB header: membrane proteinChain: C: PDB Molecule: ph stomatin;PDBTitle: crystal structure of a core domain of stomatin from2 pyrococcus horikoshii
3 c2rpbA_
99.7
16
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical membrane protein;PDBTitle: the solution structure of membrane protein
4 c2zv4O_
99.3
10
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
5 d2axtj1
78.4
24
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like6 c2kncA_
74.5
16
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
7 c1y4cA_
72.0
13
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
8 c2k1aA_
70.6
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
9 c2jwaA_
54.1
19
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
10 c3peuB_
39.6
18
PDB header: hydrolaseChain: B: PDB Molecule: nucleoporin gle1;PDBTitle: s. cerevisiae dbp5 l327v c-terminal domain bound to gle1 h337r and ip6
11 c3mk7F_
32.4
6
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
12 c1xmeB_
32.1
5
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: structure of recombinant cytochrome ba3 oxidase from thermus2 thermophilus
13 c3dktD_
31.1
16
PDB header: structural protein/virus like particleChain: D: PDB Molecule: maritimacin;PDBTitle: crystal structure of thermotoga maritima encapsulin
14 c3hd7A_
30.3
20
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
15 c2e0zA_
25.4
24
PDB header: virus like particleChain: A: PDB Molecule: virus-like particle;PDBTitle: crystal structure of virus-like particle from pyrococcus2 furiosus
16 d2axth1
21.3
23
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like17 c3hfxA_
18.4
20
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
18 c2w8aC_
17.5
23
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
19 c1cn3F_
17.4
33
PDB header: viral proteinChain: F: PDB Molecule: fragment of coat protein vp2;PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
20 c2kncB_
15.9
12
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
21 c3kdpG_
not modelled
13.8
22
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
22 c3kdpH_
not modelled
13.8
22
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
23 c3o2qB_
not modelled
11.7
29
PDB header: hydrolaseChain: B: PDB Molecule: rna polymerase ii subunit a c-terminal domain phosphatasePDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
24 c1h4uA_
not modelled
10.9
10
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
25 c3fdfA_
not modelled
10.8
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: fr253;PDBTitle: crystal structure of the serine phosphatase of rna2 polymerase ii ctd (ssu72 superfamily) from drosophila3 melanogaster. orthorhombic crystal form. northeast4 structural genomics consortium target fr253.
26 c3o2sB_
not modelled
9.9
29
PDB header: hydrolaseChain: B: PDB Molecule: rna polymerase ii subunit a c-terminal domain phosphatasePDBTitle: crystal structure of the human symplekin-ssu72 complex
27 c1zzaA_
not modelled
9.6
15
PDB header: membrane proteinChain: A: PDB Molecule: stannin;PDBTitle: solution nmr structure of the membrane protein stannin
28 c3mk7B_
not modelled
9.3
6
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit o;PDBTitle: the structure of cbb3 cytochrome oxidase
29 d2ix0a3
not modelled
8.7
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like30 c1iijA_
not modelled
8.2
13
PDB header: signaling proteinChain: A: PDB Molecule: erbb-2 receptor protein-tyrosine kinase;PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
31 c2jo1A_
not modelled
8.0
12
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
32 c1fftG_
not modelled
7.7
15
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
33 c2l2tA_
not modelled
7.6
4
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
34 d2o4ta1
not modelled
7.5
11
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like35 d2pila_
not modelled
7.2
18
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin36 d1oqwa_
not modelled
7.2
27
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin37 c3fcmA_
not modelled
7.1
7
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase, nudix family;PDBTitle: crystal structure of a nudix hydrolase from clostridium2 perfringens
38 c3ipdB_
not modelled
7.0
26
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
39 c3sokB_
not modelled
7.0
23
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
40 c2jp3A_
not modelled
6.8
6
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
41 c3a0bL_
not modelled
6.8
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
42 d1jb0i_
not modelled
6.7
22
Fold: Single transmembrane helixSuperfamily: Subunit VIII of photosystem I reaction centre, PsaIFamily: Subunit VIII of photosystem I reaction centre, PsaI43 d2r6gf1
not modelled
6.7
13
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like44 c2j7aC_
not modelled
6.6
9
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c quinol dehydrogenase nrfh;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
45 c1s5ll_
not modelled
6.5
5
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
46 c3a0hl_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
47 c3a0hL_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
48 c1s5lL_
not modelled
6.5
5
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
49 c3kziL_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
50 c2axtL_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
51 c2axtl_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
52 c3arcL_
not modelled
6.5
5
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
53 c3bz2L_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
54 c3prrL_
not modelled
6.5
5
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
55 c3a0bl_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
56 c3prqL_
not modelled
6.5
5
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
57 d2axtl1
not modelled
6.5
5
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like58 c3bz1L_
not modelled
6.5
5
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
59 d1fftb2
not modelled
6.5
6
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region60 c2zxeG_
not modelled
6.2
35
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
61 c1p58F_
not modelled
5.8
21
PDB header: virusChain: F: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
62 d1id3b_
not modelled
5.7
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones63 c2btwA_
not modelled
5.7
17
PDB header: transferaseChain: A: PDB Molecule: alr0975 protein;PDBTitle: crystal structure of alr0975
64 c3arcl_
not modelled
5.7
5
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
65 c2l16A_
not modelled
5.7
30
PDB header: protein transportChain: A: PDB Molecule: sec-independent protein translocase protein tatad;PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
66 c1yewI_
not modelled
5.6
10
PDB header: oxidoreductase, membrane proteinChain: I: PDB Molecule: particulate methane monooxygenase, b subunit;PDBTitle: crystal structure of particulate methane monooxygenase
67 c3rgbA_
not modelled
5.6
10
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase subunit b2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
68 d1uxca_
not modelled
5.6
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator69 d1j0ha1
not modelled
5.6
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes70 d2cnza1
not modelled
5.5
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits71 c2ks1B_
not modelled
5.5
15
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
72 d1jvaa3
not modelled
5.4
21
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease73 d2bu3a1
not modelled
5.4
17
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Phytochelatin synthase74 d3dtub2
not modelled
5.3
3
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region75 c2wtoB_
not modelled
5.3
9
PDB header: metal binding proteinChain: B: PDB Molecule: orf131 protein;PDBTitle: crystal structure of apo-form czce from c. metallidurans ch34
76 c3cf6E_
not modelled
5.3
17
PDB header: signaling protein/gtp-binding proteinChain: E: PDB Molecule: rap guanine nucleotide exchange factor (gef) 4;PDBTitle: structure of epac2 in complex with cyclic-amp and rap
77 c3sbtB_
not modelled
5.3
20
PDB header: splicingChain: B: PDB Molecule: a1 cistron-splicing factor aar2;PDBTitle: crystal structure of a aar2-prp8 complex
78 d2ix0a1
not modelled
5.2
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like79 c3f6aA_
not modelled
5.1
14
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase, nudix family;PDBTitle: crystal structure of a hydrolase, nudix family from2 clostridium perfringens
80 d2fhza1
not modelled
5.1
33
Fold: ImmE5-likeSuperfamily: ImmE5-likeFamily: ImmE5-like81 c2kluA_
not modelled
5.0
15
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
82 c1bhbA_
not modelled
5.0
12
PDB header: photoreceptorChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
83 d3ehbb2
not modelled
5.0
8
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region