| 1 | c3ntaA_
|
|
 |
100.0 |
25 |
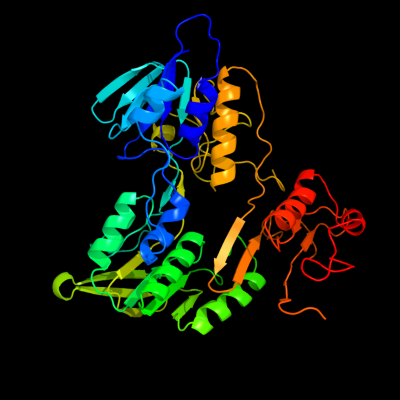
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: structure of the shewanella loihica pv-4 nadh-dependent persulfide2 reductase
|
| 2 | c3icrA_
|
|
 |
100.0 |
23 |
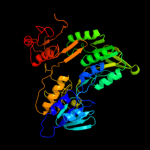
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme a-disulfide reductase;
PDBTitle: crystal structure of oxidized bacillus anthracis coadr-rhd
|
| 3 | c1zj8B_
|
|
 |
100.0 |
19 |
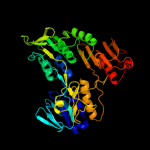
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable ferredoxin-dependent nitrite reductase nira;
PDBTitle: structure of mycobacterium tuberculosis nira protein
|
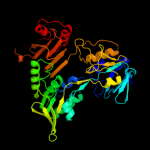
| 4 | c1gv4A_
|
|
 |
100.0 |
21 |
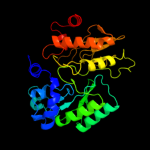
PDB header:oxidoreductase
Chain: A: PDB Molecule:programed cell death protein 8;
PDBTitle: murine apoptosis-inducing factor (aif)
|
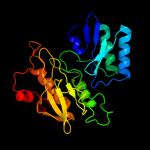
| 5 | c2akjA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin--nitrite reductase, chloroplast;
PDBTitle: structure of spinach nitrite reductase
|
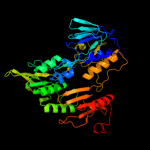
| 6 | c2gr2A_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
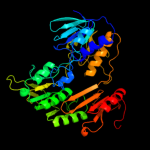
| 7 | c3lxdA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: crystal structure of ferredoxin reductase arr from novosphingobium2 aromaticivorans
|
| 8 | c1q1wA_
|
|
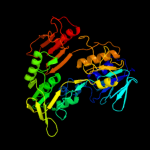 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putidaredoxin reductase;
PDBTitle: crystal structure of putidaredoxin reductase from2 pseudomonas putida
|
| 9 | c2v3aA_
|
|
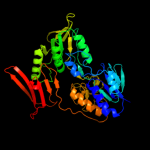 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin reductase;
PDBTitle: crystal structure of rubredoxin reductase from pseudomonas2 aeruginosa.
|
| 10 | c3oc4A_
|
|
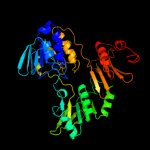 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, pyridine nucleotide-disulfide family;
PDBTitle: crystal structure of a pyridine nucleotide-disulfide family2 oxidoreductase from the enterococcus faecalis v583
|
| 11 | c1yqzA_
|
|
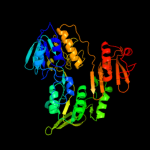 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: structure of coenzyme a-disulfide reductase from2 staphylococcus aureus refined at 1.54 angstrom resolution
|
| 12 | c3fg2P_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: P: PDB Molecule:putative rubredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase for the cyp199a2 system from2 rhodopseudomonas palustris
|
| 13 | c3iwaA_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: crystal structure of a fad-dependent pyridine nucleotide-disulphide2 oxidoreductase from desulfovibrio vulgaris
|
| 14 | c2v4jE_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 15 | c3kd9B_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: crystal structure of pyridine nucleotide disulfide oxidoreductase from2 pyrococcus horikoshii
|
| 16 | c3ef6A_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:toluene 1,2-dioxygenase system ferredoxin--nad(+)
PDBTitle: crystal structure of toluene 2,3-dioxygenase reductase
|
| 17 | c2bcpA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidase;
PDBTitle: structural analysis of streptococcus pyogenes nadh oxidase:2 c44s nox with azide
|
| 18 | c1m6iA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:programmed cell death protein 8;
PDBTitle: crystal structure of apoptosis inducing factor (aif)
|
| 19 | c3c7bE_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit beta;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 20 | c3c7bA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit alpha;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 21 | c3kljA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-dependent dehydrogenase, nirb-family (n-terminal
PDBTitle: crystal structure of nadh:rubredoxin oxidoreductase from clostridium2 acetobutylicum
|
| 22 | c1nhqA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase (h2o2(a))
Chain: A: PDB Molecule:nadh peroxidase;
PDBTitle: crystallographic analyses of nadh peroxidase cys42ala and cys42ser2 mutants: active site structure, mechanistic implications, and an3 unusual environment of arg303
|
| 23 | c2v4jA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 24 | c2cduB_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph oxidase;
PDBTitle: the crystal structure of water-forming nad(p)h oxidase from2 lactobacillus sanfranciscensis
|
| 25 | c3cgdB_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyridine nucleotide-disulfide oxidoreductase, class i;
PDBTitle: pyridine nucleotide complexes with bacillus anthracis coenzyme a-2 disulfide reductase: a structural analysis of dual nad(p)h3 specificity
|
| 26 | c1x31A_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sarcosine oxidase alpha subunit;
PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
|
| 27 | c1y56A_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1363;
PDBTitle: crystal structure of l-proline dehydrogenase from p.horikoshii
|
| 28 | c2v6oA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin glutathione reductase;
PDBTitle: structure of schistosoma mansoni thioredoxin-glutathione2 reductase (smtgr)
|
| 29 | c5aopA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase hemoprotein;
PDBTitle: sulfite reductase structure reduced with crii edta, 5-coordinate2 siroheme, siroheme feii, [4fe-4s] +1
|
| 30 | c1xhcA_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidase /nitrite reductase;
PDBTitle: nadh oxidase /nitrite reductase from pyrococcus furiosus pfu-1140779-2 001
|
| 31 | c1tytA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trypanothione reductase, oxidized form;
PDBTitle: crystal and molecular structure of crithidia fasciculata2 trypanothione reductase at 2.6 angstroms resolution
|
| 32 | c1xdiA_ |
|
not modelled |
100.0 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:rv3303c-lpda;
PDBTitle: crystal structure of lpda (rv3303c) from mycobacterium tuberculosis
|
| 33 | c2qaeA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure analysis of trypanosoma cruzi lipoamide2 dehydrogenase
|
| 34 | c2eq7B_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxoglutarate dehydrogenase e3 component;
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdo
|
| 35 | c1ojtA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:surface protein;
PDBTitle: structure of dihydrolipoamide dehydrogenase
|
| 36 | c2c3dB_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxopropyl-com reductase;
PDBTitle: 2.15 angstrom crystal structure of 2-ketopropyl coenzyme m2 oxidoreductase carboxylase with a coenzyme m disulfide3 bound at the active site
|
| 37 | c2eq8E_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
| 38 | c1lvlA_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: the refined structure of pseudomonas putida lipoamide dehydrogenase2 complexed with nad+ at 2.45 angstroms resolution
|
| 39 | c1zmcG_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of human dihydrolipoamide dehydrogenase2 complexed to nad+
|
| 40 | c1ebdB_ |
|
not modelled |
100.0 |
18 |
PDB header:complex (oxidoreductase/transferase)
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: dihydrolipoamide dehydrogenase complexed with the binding2 domain of the dihydrolipoamide acetylase
|
| 41 | c1bwcA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (glutathione reductase);
PDBTitle: structure of human glutathione reductase complexed with ajoene2 inhibitor and subversive substrate
|
| 42 | c3urhB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoamide dehydrogenase from2 sinorhizobium meliloti 1021
|
| 43 | c3ic9D_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: the structure of dihydrolipoamide dehydrogenase from colwellia2 psychrerythraea 34h.
|
| 44 | c1lpfB_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: three-dimensional structure of lipoamide dehydrogenase from2 pseudomonas fluorescens at 2.8 angstroms resolution.3 analysis of redox and thermostability properties
|
| 45 | c1zx9A_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mercuric reductase;
PDBTitle: crystal structure of tn501 mera
|
| 46 | c2a8xA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of lipoamide dehydrogenase from2 mycobacterium tuberculosis
|
| 47 | c2w0hA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trypanothione reductase;
PDBTitle: x ray structure of leishmania infantum trypanothione2 reductase in complex with antimony and nadph
|
| 48 | c1dxlC_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: dihydrolipoamide dehydrogenase of glycine decarboxylase2 from pisum sativum
|
| 49 | c1v59B_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: crystal structure of yeast lipoamide dehydrogenase2 complexed with nad+
|
| 50 | c1geuA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase(flavoenzyme)
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: anatomy of an engineered nad-binding site
|
| 51 | c3kpgA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfide-quinone reductase, putative;
PDBTitle: crystal structure of sulfide:quinone oxidoreductase from2 acidithiobacillus ferrooxidans in complex with decylubiquinone
|
| 52 | c1ndaD_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:trypanothione oxidoreductase;
PDBTitle: the structure of trypanosoma cruzi trypanothione reductase2 in the oxidized and nadph reduced state
|
| 53 | c1fcdB_ |
|
not modelled |
100.0 |
15 |
PDB header:electron transport(flavocytochrome)
Chain: B: PDB Molecule:flavocytochrome c sulfide dehydrogenase (flavin-
PDBTitle: the structure of flavocytochrome c sulfide dehydrogenase2 from a purple phototrophic bacterium chromatium vinosum at3 2.5 angstroms resolution
|
| 54 | c1hyuA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: crystal structure of intact ahpf
|
| 55 | c3dgzA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2;
PDBTitle: crystal structure of mouse mitochondrial thioredoxin reductase, c-2 terminal 3-residue truncation
|
| 56 | c2hqmB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione reductase;
PDBTitle: crystal structure of glutathione reductase glr1 from the yeast2 saccharomyces cerevisiae
|
| 57 | c2cfyB_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 1;
PDBTitle: crystal structure of human thioredoxin reductase 1
|
| 58 | c2r9zB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione amide reductase;
PDBTitle: glutathione amide reductase from chromatium gracile
|
| 59 | c3l8kB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoyl dehydrogenase from2 sulfolobus solfataricus
|
| 60 | c1zkqA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2, mitochondrial;
PDBTitle: crystal structure of mouse thioredoxin reductase type 2
|
| 61 | c1onfA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: crystal structure of plasmodium falciparum glutathione reductase
|
| 62 | c2nvkX_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of thioredoxin reductase from drosophila2 melanogaster
|
| 63 | c3o0hA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: crystal structure of glutathione reductase from bartonella henselae
|
| 64 | c3hyxC_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:sulfide-quinone reductase;
PDBTitle: 3-d x-ray structure of the sulfide:quinone oxidoreductase from aquifex2 aeolicus in complex with aurachin c
|
| 65 | c1gthD_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
|
| 66 | d1zj8a4 |
|
not modelled |
100.0 |
19 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 67 | d1aopa3 |
|
not modelled |
100.0 |
18 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 68 | d2akja4 |
|
not modelled |
100.0 |
15 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 69 | c3k30B_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 70 | d2v4jb3 |
|
not modelled |
100.0 |
19 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 71 | d3c7ba3 |
|
not modelled |
100.0 |
19 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 72 | c1ps9A_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 73 | d2v4ja3
|
|
 |
100.0 |
17 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 74 | c1djnB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 75 | d3c7bb3 |
|
not modelled |
100.0 |
17 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 76 | c3d1cA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin-containing putative monooxygenase;
PDBTitle: crystal structure of flavin-containing putative monooxygenase2 (np_373108.1) from staphylococcus aureus mu50 at 2.40 a resolution
|
| 77 | c3h8lA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidase;
PDBTitle: the first x-ray structure of a sulfide:quinone2 oxidoreductase: insights into sulfide oxidation mechanism
|
| 78 | c2zbwA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of thioredoxin reductase-like protein from thermus2 thermophilus hb8
|
| 79 | d1zj8a3 |
|
not modelled |
100.0 |
18 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 80 | c2vdcI_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glutamate synthase [nadph] small chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 81 | c3r9uA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: thioredoxin-disulfide reductase from campylobacter jejuni.
|
| 82 | c3f8rD_ |
|
not modelled |
99.9 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:thioredoxin reductase (trxb-3);
PDBTitle: crystal structure of sulfolobus solfataricus thioredoxin2 reductase b3 in complex with two nadp molecules
|
| 83 | c1vdcA_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dependent thioredoxin reductase;
PDBTitle: structure of nadph dependent thioredoxin reductase
|
| 84 | c3ctyA_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of t. acidophilum thioredoxin reductase
|
| 85 | c1lqtB_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fpra;
PDBTitle: a covalent modification of nadp+ revealed by the atomic resolution2 structure of fpra, a mycobacterium tuberculosis oxidoreductase
|
| 86 | c1cjcA_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (adrenodoxin reductase);
PDBTitle: structure of adrenodoxin reductase of mitochondrial p4502 systems
|
| 87 | c3ab1B_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin--nadp reductase;
PDBTitle: crystal structure of ferredoxin nadp+ oxidoreductase
|
| 88 | c1fl2A_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: catalytic core component of the alkylhydroperoxide reductase ahpf from2 e.coli
|
| 89 | c3d8xB_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 1;
PDBTitle: crystal structure of saccharomyces cerevisiae nadph dependent2 thioredoxin reductase 1
|
| 90 | d1aopa4 |
|
not modelled |
99.9 |
14 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 91 | c3s5wB_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-ornithine 5-monooxygenase;
PDBTitle: ornithine hydroxylase (pvda) from pseudomonas aeruginosa
|
| 92 | c3lzxB_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin--nadp reductase 2;
PDBTitle: crystal structure of ferredoxin-nadp+ oxidoreductase from bacillus2 subtilis (form ii)
|
| 93 | c1f6mF_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of a complex between thioredoxin2 reductase, thioredoxin, and the nadp+ analog, aadp+
|
| 94 | c3fbsB_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of the oxidoreductase from agrobacterium2 tumefaciens
|
| 95 | c2a87A_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of m. tuberculosis thioredoxin reductase
|
| 96 | c2q0lA_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: helicobacter pylori thioredoxin reductase reduced by sodium dithionite2 in complex with nadp+
|
| 97 | c2vq7B_ |
|
not modelled |
99.9 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:flavin-containing monooxygenase;
PDBTitle: bacterial flavin-containing monooxygenase in complex with2 nadp: native data
|
| 98 | d2akja3 |
|
not modelled |
99.9 |
15 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 99 | c2q7vA_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of deinococcus radiodurans thioredoxin2 reductase
|
| 100 | c3gwdA_ |
|
not modelled |
99.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cyclohexanone monooxygenase;
PDBTitle: closed crystal structure of cyclohexanone monooxygenase
|
| 101 | c1w4xA_ |
|
not modelled |
99.9 |
21 |
PDB header:oxygenase
Chain: A: PDB Molecule:phenylacetone monooxygenase;
PDBTitle: phenylacetone monooxygenase, a baeyer-villiger2 monooxygenase
|
| 102 | c1vqwB_ |
|
not modelled |
99.9 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein with similarity to flavin-containing
PDBTitle: crystal structure of a protein with similarity to flavin-2 containing monooxygenases and to mammalian dimethylalanine3 monooxygenases
|
| 103 | d1zj8a2 |
|
not modelled |
99.9 |
17 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 104 | d1gv4a1 |
|
not modelled |
99.8 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 105 | d1d7ya1 |
|
not modelled |
99.8 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 106 | d1q1ra1 |
|
not modelled |
99.8 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 107 | d1xhca1 |
|
not modelled |
99.8 |
25 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 108 | c2rghA_ |
|
not modelled |
99.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 109 | d1m6ia1 |
|
not modelled |
99.8 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 110 | d1nhpa1 |
|
not modelled |
99.8 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 111 | c3da1A_ |
|
not modelled |
99.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
|
| 112 | c2rgoA_ |
|
not modelled |
99.7 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 113 | d1d7ya2 |
|
not modelled |
99.7 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 114 | d3grsa1 |
|
not modelled |
99.7 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 115 | d1xdia1 |
|
not modelled |
99.7 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 116 | d1mo9a1 |
|
not modelled |
99.7 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 117 | d2akja2 |
|
not modelled |
99.7 |
16 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 118 | d1nhpa2 |
|
not modelled |
99.7 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 119 | d1q1ra2 |
|
not modelled |
99.7 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 120 | d1fcda1 |
|
not modelled |
99.7 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |