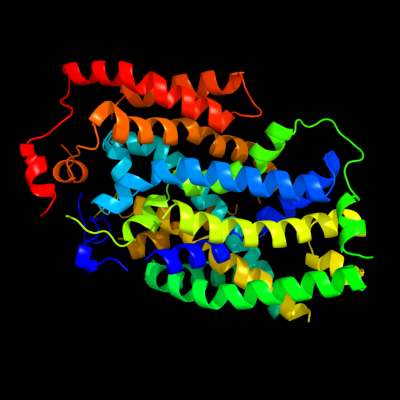
1 c2jlnA_
100.0
20
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
2 c2xq2A_
99.4
14
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
3 c3giaA_
99.3
13
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
4 c3dh4A_
99.2
16
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
5 c3lrcC_
98.8
11
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
6 c3hfxA_
92.9
10
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
7 d2a65a1
89.4
14
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like8 c3qnqD_
70.0
14
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
9 c2w8aC_
53.0
16
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
10 d2oara1
24.2
11
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel11 c1bh0A_
24.0
23
PDB header: synthetic hormoneChain: A: PDB Molecule: glucagon;PDBTitle: structure of a glucagon analog
12 c2ke4A_
19.8
23
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
13 c1nauA_
18.8
10
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon;PDBTitle: nmr solution structure of the glucagon antagonist [deshis1,2 desphe6, glu9]glucagon amide in the presence of3 perdeuterated dodecylphosphocholine micelles
14 c3o2tA_
18.5
18
PDB header: protein bindingChain: A: PDB Molecule: symplekin;PDBTitle: crystal structure of the n-terminal domain of human symplekin
15 c3o2qD_
17.8
20
PDB header: hydrolaseChain: D: PDB Molecule: symplekin;PDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
16 c3odrA_
17.8
20
PDB header: protein bindingChain: A: PDB Molecule: symplekin;PDBTitle: crystal structure of the n-terminal domain of human symplekin
17 c3odsA_
15.3
20
PDB header: protein bindingChain: A: PDB Molecule: symplekin;PDBTitle: crystal structure of the k185a mutant of the n-terminal domain of2 human symplekin
18 c3gs3A_
14.7
31
PDB header: transcription, protein bindingChain: A: PDB Molecule: symplekin;PDBTitle: structure of the n-terminal heat domain of symplekin from d.2 melanogaster
19 c2oarA_
13.8
11
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
20 d1t98a1
11.2
42
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MukF N-terminal domain-like21 c1d0rA_
not modelled
10.7
18
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
22 c2wj8N_
not modelled
10.7
15
PDB header: rna binding protein/rnaChain: N: PDB Molecule: nucleoprotein;PDBTitle: respiratory syncitial virus ribonucleoprotein
23 c2ntxB_
not modelled
10.7
19
PDB header: signaling proteinChain: B: PDB Molecule: emb24 d1mpga2
not modelled
10.5
13
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: DNA repair glycosylase, N-terminal domain25 d1iyjb5
not modelled
9.7
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB26 c3mtvA_
not modelled
9.7
29
PDB header: hydrolaseChain: A: PDB Molecule: papain-like cysteine protease;PDBTitle: the crystal structure of the prrsv nonstructural protein nsp1
27 d1xrsb2
not modelled
9.6
29
Fold: Dodecin subunit-likeSuperfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domainFamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain28 c2kmgA_
not modelled
9.2
20
PDB header: gene regulationChain: A: PDB Molecule: klca;PDBTitle: the structure of the klca and ardb proteins show a novel2 fold and antirestriction activity against type i dna3 restriction systems in vivo but not in vitro
29 c2bbjB_
not modelled
8.9
14
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
30 d2dnta1
not modelled
8.5
8
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: Chromo domain31 d1ci4a_
not modelled
7.8
14
Fold: SAM domain-likeSuperfamily: Barrier-to-autointegration factor, BAFFamily: Barrier-to-autointegration factor, BAF32 c3dvkB_
not modelled
7.5
30
PDB header: membrane proteinChain: B: PDB Molecule: voltage-dependent r-type calcium channel subunit alpha-1e;PDBTitle: crystal structure of ca2+/cam-cav2.3 iq domain complex
33 d3cx5e2
not modelled
7.3
18
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor34 d1miua5
not modelled
7.3
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB35 c1jrjA_
not modelled
7.0
14
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
36 c2czsB_
not modelled
6.9
19
PDB header: electron transportChain: B: PDB Molecule: cytochrome c, putative;PDBTitle: crystal structure analysis of the diheme c-type cytochrome dhc2
37 c3c66B_
not modelled
6.7
14
PDB header: transferaseChain: B: PDB Molecule: poly(a) polymerase;PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
38 c2lf6A_
not modelled
6.7
33
PDB header: signaling proteinChain: A: PDB Molecule: effector protein hopab1;PDBTitle: solution nmr structure of hopabpph1448_220_320 from pseudomonas2 syringae pv. phaseolicola str. 1448a, midwest center for structural3 genomics target apc40132.4 and northeast structural genomics4 consortium target pst3a
39 d1lgha_
not modelled
6.6
40
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits40 d1fcda2
not modelled
6.5
10
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains41 c3siqF_
not modelled
6.4
20
PDB header: ligaseChain: F: PDB Molecule: apoptosis 1 inhibitor;PDBTitle: crystal structure of autoinhibited diap1-bir1 domain
42 d1jj2y_
not modelled
6.4
33
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae43 c3g36D_
not modelled
6.3
21
PDB header: nuclear proteinChain: D: PDB Molecule: protein dpy-30 homolog;PDBTitle: crystal structure of the human dpy-30-like c-terminal domain
44 d1qi9a_
not modelled
6.2
25
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)45 c3ilaG_
not modelled
6.2
29
PDB header: signaling proteinChain: G: PDB Molecule: ryanodine receptor 1;PDBTitle: crystal structure of rabbit ryanodine receptor 1 n-terminal domain (9-2 205)
46 c1yshD_
not modelled
6.2
33
PDB header: structural protein/rnaChain: D: PDB Molecule: ribosomal protein l37a;PDBTitle: localization and dynamic behavior of ribosomal protein l30e
47 d1v4sa1
not modelled
6.1
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase48 c4a17Y_
not modelled
6.1
17
PDB header: ribosomeChain: Y: PDB Molecule: rpl37a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
49 c2kvlA_
not modelled
6.1
30
PDB header: viral proteinChain: A: PDB Molecule: major outer capsid protein vp7;PDBTitle: nmr structure of the c-terminal domain of vp7
50 d1bcce2
not modelled
6.1
13
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor51 c2ht2B_
not modelled
6.0
14
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
52 c2lf3A_
not modelled
5.9
17
PDB header: signaling proteinChain: A: PDB Molecule: effector protein hopab3;PDBTitle: solution nmr structure of hoppmal_281_385 from pseudomonas syringae2 pv. maculicola str. es4326, midwest center for structural genomics3 target apc40104.5 and northeast structural genomics consortium target4 pst2a
53 c3bxlB_
not modelled
5.9
21
PDB header: membrane protein, signaling proteinChain: B: PDB Molecule: voltage-dependent r-type calcium channel subunitPDBTitle: crystal structure of the r-type calcium channel (cav2.3) iq2 domain and ca2+calmodulin complex
54 d1rg6a_
not modelled
5.9
10
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain55 c1ciiA_
not modelled
5.9
12
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
56 c3cc4Z_
not modelled
5.8
50
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: co-crystal structure of anisomycin bound to the 50s ribosomal subunit
57 d2q66a1
not modelled
5.8
14
Fold: PAP/OAS1 substrate-binding domainSuperfamily: PAP/OAS1 substrate-binding domainFamily: Poly(A) polymerase, PAP, middle domain58 c2qa4Z_
not modelled
5.8
44
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: a more complete structure of the the l7/l12 stalk of the2 haloarcula marismortui 50s large ribosomal subunit
59 d1vqoz1
not modelled
5.8
44
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae60 d1ppje2
not modelled
5.8
27
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor61 d2o8pa1
not modelled
5.7
33
Fold: alpha-alpha superhelixSuperfamily: 14-3-3 proteinFamily: 14-3-3 protein62 d1bg3a3
not modelled
5.6
13
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase63 d3bbda1
not modelled
5.6
8
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: EMG1/NEP1-like64 d1wgma_
not modelled
5.5
7
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box65 d1bdga1
not modelled
5.5
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase66 d1a7ja_
not modelled
5.5
8
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase67 d1j72a1
not modelled
5.5
25
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like68 c2v6zM_
not modelled
5.5
7
PDB header: transferaseChain: M: PDB Molecule: dna polymerase epsilon subunit 2;PDBTitle: solution structure of amino terminal domain of human dna2 polymerase epsilon subunit b
69 d3efza1
not modelled
5.4
33
Fold: alpha-alpha superhelixSuperfamily: 14-3-3 proteinFamily: 14-3-3 protein70 c3efzA_
not modelled
5.4
33
PDB header: signaling proteinChain: A: PDB Molecule: 14-3-3 protein;PDBTitle: crystal structure of a 14-3-3 protein from cryptosporidium parvum2 (cgd1_2980)
71 c2kncA_
not modelled
5.4
28
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
72 d1ffkw_
not modelled
5.4
40
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae73 c3o7bA_
not modelled
5.4
33
PDB header: transferaseChain: A: PDB Molecule: ribosome biogenesis nep1 rna methyltransferase;PDBTitle: crystal structure of archaeoglobus fulgidus nep1 bound to s-2 adenosylhomocysteine
74 c1afoB_
not modelled
5.4
17
PDB header: integral membrane proteinChain: B: PDB Molecule: glycophorin a;PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
75 c2voyB_
not modelled
5.3
9
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
76 c2w9jB_
not modelled
5.2
10
PDB header: signaling proteinChain: B: PDB Molecule: signal recognition particle subunit srp14;PDBTitle: the crystal structure of srp14 from the schizosaccharomyces2 pombe signal recognition particle
77 c2rowA_
not modelled
5.2
17
PDB header: transferaseChain: A: PDB Molecule: rho-associated protein kinase 2;PDBTitle: the c1 domain of rock ii
78 c2kb1A_
not modelled
5.2
16
PDB header: membrane proteinChain: A: PDB Molecule: wsk3;PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
79 d1otsa_
not modelled
5.2
14
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel80 c1z65A_
not modelled
5.1
15
PDB header: unknown functionChain: A: PDB Molecule: prion-like protein doppel;PDBTitle: mouse doppel 1-30 peptide
81 c3jyw9_
not modelled
5.1
38
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
82 d2iuba2
not modelled
5.0
5
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region83 c3dvmB_
not modelled
5.0
13
PDB header: membrane proteinChain: B: PDB Molecule: voltage-dependent p/q-type calcium channel subunit alpha-PDBTitle: crystal structure of ca2+/cam-cav2.1 iq domain complex
84 d2jn4a1
not modelled
5.0
0
Fold: NifT/FixU barrel-likeSuperfamily: NifT/FixU-likeFamily: NifT/FixU