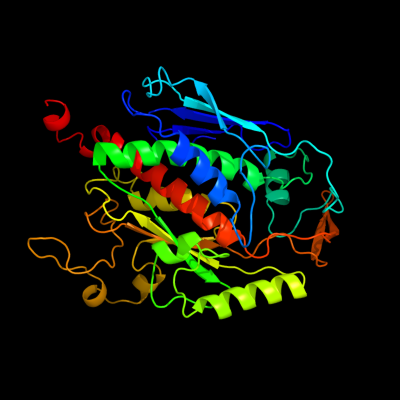
| 1 | d1um0a_
|
|
 |
100.0 |
45 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
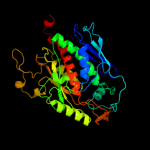
| 2 | d1qxoa_
|
|
 |
100.0 |
38 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
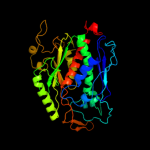
| 3 | c1ztbA_
|
|
 |
100.0 |
38 |
PDB header:ligase
Chain: A: PDB Molecule:chorismate synthase;
PDBTitle: crystal structure of chorismate synthase from mycobacterium2 tuberculosis
|
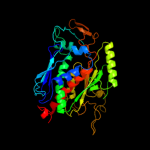
| 4 | d1q1la_
|
|
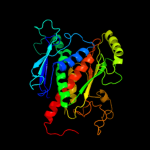 |
100.0 |
41 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
| 5 | d1sq1a_
|
|
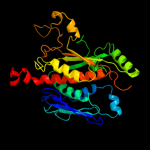 |
100.0 |
45 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
| 6 | d1r53a_
|
|
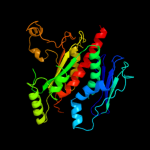 |
100.0 |
51 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
| 7 | c2k1hA_
|
|
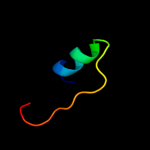 |
76.4 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ser13;
PDBTitle: solution nmr structure of ser13 from staphylococcus epidermidis.2 northeast structural genomics consortium target ser13
|
| 8 | d2ffma1
|
|
 |
73.0 |
13 |
Fold:Hypothetical protein SAV1430
Superfamily:Hypothetical protein SAV1430
Family:Hypothetical protein SAV1430 |
| 9 | c3r3sD_
|
|
 |
61.2 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase;
PDBTitle: structure of the ygha oxidoreductase from salmonella enterica
|
| 10 | d1o4va_
|
|
 |
61.1 |
25 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 11 | c3lp6D_
|
|
 |
58.6 |
31 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
| 12 | d1qcza_
|
|
 |
54.8 |
21 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 13 | c1m1gB_
|
|
 |
53.4 |
28 |
PDB header:transcription
Chain: B: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
|
| 14 | d1xmpa_
|
|
 |
44.9 |
23 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 15 | c2ekmC_
|
|
 |
40.6 |
24 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein st1511;
PDBTitle: structure of st1219 protein from sulfolobus tokodaii
|
| 16 | d1vgga_
|
|
 |
39.1 |
23 |
Fold:Ta1353-like
Superfamily:Ta1353-like
Family:Ta1353-like |
| 17 | c3trhI_
|
|
 |
35.4 |
24 |
PDB header:lyase
Chain: I: PDB Molecule:phosphoribosylaminoimidazole carboxylase
PDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
|
| 18 | c2xznF_
|
|
 |
35.4 |
13 |
PDB header:ribosome
Chain: F: PDB Molecule:eif1;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 19 | c2lhuA_
|
|
 |
32.7 |
27 |
PDB header:structural protein
Chain: A: PDB Molecule:mybpc3 protein;
PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
|
| 20 | d1ciya1
|
|
 |
31.1 |
33 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:delta-Endotoxin, C-terminal domain |
| 21 | d1ji6a1 |
|
not modelled |
29.3 |
24 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:delta-Endotoxin, C-terminal domain |
| 22 | c2d16B_ |
|
not modelled |
27.9 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph1918;
PDBTitle: crystal structure of ph1918 protein from pyrococcus horikoshii ot3
|
| 23 | c3orsD_ |
|
not modelled |
26.5 |
16 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
| 24 | c3rggD_ |
|
not modelled |
23.0 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
|
| 25 | d1dlca1 |
|
not modelled |
22.6 |
29 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:delta-Endotoxin, C-terminal domain |
| 26 | d1qlwa_ |
|
not modelled |
22.6 |
28 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:A novel bacterial esterase |
| 27 | c2ywxA_ |
|
not modelled |
17.8 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
|
| 28 | d2za7a1 |
|
not modelled |
16.0 |
42 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
| 29 | d1z6om1 |
|
not modelled |
15.9 |
32 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
| 30 | c2w3pB_ |
|
not modelled |
13.9 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:benzoyl-coa-dihydrodiol lyase;
PDBTitle: boxc crystal structure
|
| 31 | c2ejeA_ |
|
not modelled |
13.7 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i;
PDBTitle: solution structure of rsgi ruh-071, a gtf2i domain in human2 cdna
|
| 32 | c3mcaB_ |
|
not modelled |
13.1 |
20 |
PDB header:translation regulation/hydrolase
Chain: B: PDB Molecule:protein dom34;
PDBTitle: structure of the dom34-hbs1 complex and implications for its role in2 no-go decay
|
| 33 | c2dn5A_ |
|
not modelled |
13.0 |
42 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i repeat domain-
PDBTitle: solution structure of rsgi ruh-057, a gtf2i domain in human2 cdna
|
| 34 | c2a73B_ |
|
not modelled |
12.7 |
16 |
PDB header:immune system
Chain: B: PDB Molecule:complement c3;
PDBTitle: human complement component c3
|
| 35 | c2dzrA_ |
|
not modelled |
12.3 |
58 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i repeat domain-
PDBTitle: solution structure of rsgi ruh-067, a gtf2i domain in human2 cdna
|
| 36 | d1q60a_ |
|
not modelled |
12.1 |
42 |
Fold:GTF2I-like repeat
Superfamily:GTF2I-like repeat
Family:GTF2I-like repeat |
| 37 | c2d99A_ |
|
not modelled |
12.1 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i repeat domain-
PDBTitle: solution structure of rsgi ruh-048, a gtf2i domain in human2 cdna
|
| 38 | c1w99A_ |
|
not modelled |
12.1 |
24 |
PDB header:toxin
Chain: A: PDB Molecule:pesticidial crystal protein cry4ba;
PDBTitle: mosquito-larvicidal toxin cry4ba from bacillus2 thuringiensis ssp. israelensis
|
| 39 | c3b8hA_ |
|
not modelled |
11.9 |
50 |
PDB header:biosynthetic protein/transferase
Chain: A: PDB Molecule:elongation factor 2;
PDBTitle: structure of the eef2-exoa(e546a)-nad+ complex
|
| 40 | c2dn4A_ |
|
not modelled |
11.7 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i;
PDBTitle: solution structure of rsgi ruh-060, a gtf2i domain in human2 cdna
|
| 41 | d1r03a_ |
|
not modelled |
11.4 |
43 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
| 42 | d1g9pa_ |
|
not modelled |
11.3 |
100 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:omega toxin-like
Family:Spider toxins |
| 43 | c1g9pA_ |
|
not modelled |
11.3 |
100 |
PDB header:toxin
Chain: A: PDB Molecule:omega-atracotoxin-hv2a;
PDBTitle: solution structure of the insecticidal calcium channel2 blocker omega-atracotoxin-hv2a
|
| 44 | c2c9kA_ |
|
not modelled |
11.1 |
38 |
PDB header:toxin
Chain: A: PDB Molecule:pesticidal crystal protein cry4aa;
PDBTitle: structure of the functional form of the mosquito-larvicidal2 cry4aa toxin from bacillus thuringiensis at 2.8 a3 resolution
|
| 45 | c2k4mA_ |
|
not modelled |
11.1 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein2 mth_1000, northeast structural genomics consortium target3 tr8
|
| 46 | c2e3lA_ |
|
not modelled |
10.8 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:transcription factor gtf2ird2 beta;
PDBTitle: solution structure of rsgi ruh-068, a gtf2i domain in human2 cdna
|
| 47 | c2ed2A_ |
|
not modelled |
10.7 |
42 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i;
PDBTitle: solution structure of rsgi ruh-069, a gtf2i domain in human2 cdna
|
| 48 | d2ceia1 |
|
not modelled |
10.5 |
40 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
| 49 | c2dzqA_ |
|
not modelled |
10.4 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:general transcription factor ii-i repeat domain-
PDBTitle: solution structure of rsgi ruh-066, a gtf2i domain in human2 cdna
|
| 50 | d1rcda_ |
|
not modelled |
10.0 |
32 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
| 51 | d1x52a1 |
|
not modelled |
9.7 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
| 52 | d2fzcb1 |
|
not modelled |
9.3 |
30 |
Fold:Ferredoxin-like
Superfamily:Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain
Family:Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain |
| 53 | c2h3eB_ |
|
not modelled |
8.7 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase regulatory chain;
PDBTitle: structure of wild-type e. coli aspartate transcarbamoylase in the2 presence of n-phosphonacetyl-l-isoasparagine at 2.3a resolution
|
| 54 | d1pg5b1 |
|
not modelled |
8.6 |
40 |
Fold:Ferredoxin-like
Superfamily:Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain
Family:Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain |
| 55 | d1nz9a_ |
|
not modelled |
8.6 |
53 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
| 56 | c2b39B_ |
|
not modelled |
8.5 |
17 |
PDB header:immune system
Chain: B: PDB Molecule:c3;
PDBTitle: structure of mammalian c3 with an intact thioester at 3a resolution
|
| 57 | c3izq1_ |
|
not modelled |
8.3 |
33 |
PDB header:ribosomal protein,hydrolase
Chain: 1: PDB Molecule:elongation factor 1 alpha-like protein;
PDBTitle: structure of the dom34-hbs1-gdpnp complex bound to a translating2 ribosome
|
| 58 | c3degC_ |
|
not modelled |
8.1 |
26 |
PDB header:ribosome
Chain: C: PDB Molecule:gtp-binding protein lepa;
PDBTitle: complex of elongating escherichia coli 70s ribosome and ef4(lepa)-2 gmppnp
|
| 59 | d2do3a1 |
|
not modelled |
8.1 |
47 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:SPT5 KOW domain-like |
| 60 | c2be7E_ |
|
not modelled |
7.9 |
15 |
PDB header:transferase
Chain: E: PDB Molecule:aspartate carbamoyltransferase regulatory chain;
PDBTitle: crystal structure of the unliganded (t-state) aspartate2 transcarbamoylase of the psychrophilic bacterium moritella profunda
|
| 61 | c1pg5B_ |
|
not modelled |
7.7 |
40 |
PDB header:transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase regulatory chain;
PDBTitle: crystal structure of the unligated (t-state) aspartate2 transcarbamoylase from the extremely thermophilic archaeon sulfolobus3 acidocaldarius
|
| 62 | c2oghA_ |
|
not modelled |
7.7 |
13 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor eif-1;
PDBTitle: solution structure of yeast eif1
|
| 63 | c2ywfA_ |
|
not modelled |
7.6 |
24 |
PDB header:translation
Chain: A: PDB Molecule:gtp-binding protein lepa;
PDBTitle: crystal structure of gmppnp-bound lepa from aquifex aeolicus
|
| 64 | c3bdeA_ |
|
not modelled |
7.2 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:mll5499 protein;
PDBTitle: crystal structure of a dabb family protein with a ferredoxin-like fold2 (mll5499) from mesorhizobium loti maff303099 at 1.79 a resolution
|
| 65 | c2xg8D_ |
|
not modelled |
7.2 |
38 |
PDB header:transcription
Chain: D: PDB Molecule:pipx;
PDBTitle: structural basis of gene regulation by protein pii: the2 crystal complex of pii and pipx from synechococcus3 elongatus pcc 7942
|
| 66 | c3n5bB_ |
|
not modelled |
7.1 |
38 |
PDB header:transcription regulator
Chain: B: PDB Molecule:asr0485 protein;
PDBTitle: the complex of pii and pipx from anabaena
|
| 67 | d2vgna3 |
|
not modelled |
7.1 |
7 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
| 68 | c3cb4D_ |
|
not modelled |
7.1 |
31 |
PDB header:translation
Chain: D: PDB Molecule:gtp-binding protein lepa;
PDBTitle: the crystal structure of lepa
|
| 69 | c3kopB_ |
|
not modelled |
7.0 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
|
| 70 | c2diuA_ |
|
not modelled |
6.9 |
28 |
PDB header:rna binding protein
Chain: A: PDB Molecule:kiaa0430 protein;
PDBTitle: solution structure of the rrm domain of kiaa0430 protein
|
| 71 | c3seeA_ |
|
not modelled |
6.8 |
18 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:hypothetical sugar binding protein;
PDBTitle: crystal structure of a hypothetical sugar binding protein (bt_4411)2 from bacteroides thetaiotaomicron vpi-5482 at 1.25 a resolution
|
| 72 | c3ebkA_ |
|
not modelled |
6.7 |
50 |
PDB header:allergen
Chain: A: PDB Molecule:allergen bla g 4;
PDBTitle: crystal structure of major allergens, bla g 4 from2 cockroaches
|
| 73 | d1nppa2 |
|
not modelled |
6.6 |
43 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
| 74 | c2xexA_ |
|
not modelled |
6.5 |
60 |
PDB header:translation
Chain: A: PDB Molecule:elongation factor g;
PDBTitle: crystal structure of staphylococcus aureus elongation factor2 g
|
| 75 | d1l5oa_ |
|
not modelled |
6.5 |
21 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
| 76 | c3tr5C_ |
|
not modelled |
6.5 |
47 |
PDB header:translation
Chain: C: PDB Molecule:peptide chain release factor 3;
PDBTitle: structure of a peptide chain release factor 3 (prfc) from coxiella2 burnetii
|
| 77 | c2ywwA_ |
|
not modelled |
6.5 |
22 |
PDB header:metal binding protein
Chain: A: PDB Molecule:aspartate carbamoyltransferase regulatory chain;
PDBTitle: crystal structure of aspartate carbamoyltransferase2 regulatory chain from methanocaldococcus jannaschii
|
| 78 | c1ciyA_ |
|
not modelled |
6.2 |
32 |
PDB header:toxin
Chain: A: PDB Molecule:cryia(a);
PDBTitle: insecticidal toxin: structure and channel formation
|
| 79 | c1dlcA_ |
|
not modelled |
5.9 |
28 |
PDB header:toxin
Chain: A: PDB Molecule:delta-endotoxin cryiiia;
PDBTitle: crystal structure of insecticidal delta-endotoxin from2 bacillus thuringiensis at 2.5 angstroms resolution
|
| 80 | d2atcb1 |
|
not modelled |
5.9 |
20 |
Fold:Ferredoxin-like
Superfamily:Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain
Family:Aspartate carbamoyltransferase, Regulatory-chain, N-terminal domain |
| 81 | c3bb5B_ |
|
not modelled |
5.8 |
9 |
PDB header:unknown function
Chain: B: PDB Molecule:stress responsive alpha-beta protein;
PDBTitle: crystal structure of a dimeric ferredoxin-like protein of unknown2 function (jann_3925) from jannaschia sp. ccs1 at 2.30 a resolution
|
| 82 | c2zxeG_ |
|
not modelled |
5.6 |
33 |
PDB header:hydrolase/transport protein
Chain: G: PDB Molecule:phospholemman-like protein;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
|
| 83 | c1ji6A_ |
|
not modelled |
5.4 |
19 |
PDB header:toxin
Chain: A: PDB Molecule:pesticidial crystal protein cry3bb;
PDBTitle: crystal structure of the insecticidal bacterial del2 endotoxin cry3bb1 bacillus thuringiensis
|
| 84 | d1vqot1 |
|
not modelled |
5.3 |
35 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
| 85 | c1zn0B_ |
|
not modelled |
5.1 |
60 |
PDB header:translation/biosynthetic protein/rna
Chain: B: PDB Molecule:elongation factor g;
PDBTitle: coordinates of rrf and ef-g fitted into cryo-em map of the2 50s subunit bound with both ef-g (gdpnp) and rrf
|