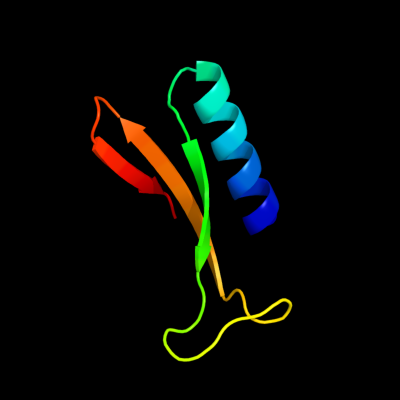
1 c3kd4A_
79.8
19
PDB header: hydrolaseChain: A: PDB Molecule: putative protease;PDBTitle: crystal structure of a putative protease (bdi_1141) from2 parabacteroides distasonis atcc 8503 at 2.00 a resolution
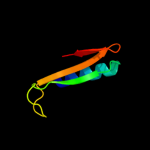
2 d2f4ma1
38.4
17
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core3 c3jubA_
37.9
14
PDB header: transferaseChain: A: PDB Molecule: aig2-like domain-containing protein 1;PDBTitle: human gamma-glutamylamine cyclotransferase
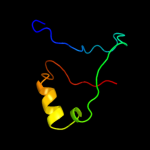
4 d1rq0a_
34.5
16
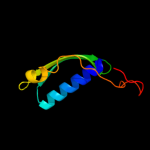
Fold: Release factorSuperfamily: Release factorFamily: Release factor5 c1zbtA_
29.2
19
PDB header: translationChain: A: PDB Molecule: peptide chain release factor 1;PDBTitle: crystal structure of peptide chain release factor 1 (rf-1) (smu.1085)2 from streptococcus mutans at 2.34 a resolution
6 d1gqea_
28.1
20
Fold: Release factorSuperfamily: Release factorFamily: Release factor7 d2b3tb1
26.2
17
Fold: Release factorSuperfamily: Release factorFamily: Release factor8 c3u7jA_
22.1
29
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
9 c3nzlA_
20.4
30
PDB header: transcriptionChain: A: PDB Molecule: dna-binding protein satb1;PDBTitle: crystal structure of the n-terminal domain of dna-binding protein2 satb1 from homo sapiens, northeast structural genomics consortium3 target hr4435b
10 c3necD_
19.8
29
PDB header: actin-binding proteinChain: D: PDB Molecule: inflammatory profilin;PDBTitle: crystal structure of toxoplasma gondii profilin
11 d1vkba_
17.7
10
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like12 c3h25A_
17.5
38
PDB header: replication/dnaChain: A: PDB Molecule: replication protein b;PDBTitle: crystal structure of the catalytic domain of primase repb' in complex2 with initiator dna
13 c2ihr1_
15.4
12
PDB header: translationChain: 1: PDB Molecule: peptide chain release factor 2;PDBTitle: rf2 of thermus thermophilus
14 c3bg4D_
13.7
40
PDB header: hydrolase/hydrolase inhibitorChain: D: PDB Molecule: guamerin;PDBTitle: the crystal structure of guamerin in complex with2 chymotrypsin and the development of an elastase-specific3 inhibitor
15 c3d5cX_
13.0
15
PDB header: ribosomeChain: X: PDB Molecule: peptide chain release factor 1;PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 30s subunit, release factor 1 (rf1), two trna, and3 mrna molecules of the second 70s ribosome. the entire crystal4 structure contains two 70s ribosomes as described in remark 400.
16 c2z8tX_
12.8
14
PDB header: hydrolaseChain: X: PDB Molecule: protein-glutaminase;PDBTitle: crystal structure of protein-glutaminase of c.proteolyticum2 strain 9670
17 c3d9wA_
12.2
14
PDB header: transferaseChain: A: PDB Molecule: putative acetyltransferase;PDBTitle: crystal structure analysis of nocardia farcinica arylamine2 n-acetyltransferase
18 c3imkA_
12.1
35
PDB header: metal binding proteinChain: A: PDB Molecule: putative molybdenum carrier protein;PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
19 d1ozha2
11.9
15
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module20 c2kp7A_
11.2
19
PDB header: hydrolaseChain: A: PDB Molecule: crossover junction endonuclease mus81;PDBTitle: solution nmr structure of the mus81 n-terminal hhh.2 northeast structural genomics consortium target mmt1a
21 d1f2ka_
not modelled
10.7
26
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)22 d1x3za1
not modelled
10.5
20
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core23 d2r4qa1
not modelled
10.1
21
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like24 d1hn6a_
not modelled
9.7
54
Fold: Apical membrane antigen 1Superfamily: Apical membrane antigen 1Family: Apical membrane antigen 125 d1pnea_
not modelled
9.3
9
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)26 d1bx7a_
not modelled
9.0
40
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Huristasin-like27 c3rq4A_
not modelled
8.8
18
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv420h2;PDBTitle: crystal structure of suppressor of variegation 4-20 homolog 2
28 d2hlya1
not modelled
8.4
20
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Atu2299-like29 d1acfa_
not modelled
8.2
26
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)30 c3pifD_
not modelled
8.1
14
PDB header: hydrolaseChain: D: PDB Molecule: 5'->3' exoribonuclease (xrn1);PDBTitle: crystal structure of the 5'->3' exoribonuclease xrn1, e178q mutant in2 complex with manganese
31 d3nula_
not modelled
8.0
32
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)32 d1cqaa_
not modelled
8.0
27
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)33 d1brza_
not modelled
8.0
19
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Plant defensins34 d1g71a_
not modelled
7.8
22
Fold: Prim-pol domainSuperfamily: Prim-pol domainFamily: PriA-like35 d2pbdp1
not modelled
7.7
9
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)36 d2c1ha1
not modelled
7.3
21
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)37 d1g5ua_
not modelled
7.3
27
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)38 d2r48a1
not modelled
7.3
26
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like39 c3davA_
not modelled
7.2
26
PDB header: protein bindingChain: A: PDB Molecule: profilin;PDBTitle: schizosaccharomyces pombe profilin crystallized from sodium2 formate
40 d1v30a_
not modelled
6.9
6
Fold: Gamma-glutamyl cyclotransferase-likeSuperfamily: Gamma-glutamyl cyclotransferase-likeFamily: Gamma-glutamyl cyclotransferase-like41 c3enpA_
not modelled
6.8
24
PDB header: hydrolaseChain: A: PDB Molecule: tp53rk-binding protein;PDBTitle: crystal structure of human cgi121
42 d1ejab_
not modelled
6.8
40
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Huristasin-like43 d1s7ja_
not modelled
6.7
12
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like44 d1ypra_
not modelled
6.6
26
Fold: Profilin-likeSuperfamily: Profilin (actin-binding protein)Family: Profilin (actin-binding protein)45 d1l6sa_
not modelled
6.0
21
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)46 d1umga_
not modelled
5.9
31
Fold: Sulfolobus fructose-1,6-bisphosphatase-likeSuperfamily: Sulfolobus fructose-1,6-bisphosphatase-likeFamily: Sulfolobus fructose-1,6-bisphosphatase-like47 d1o8bb1
not modelled
5.6
25
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain48 d2zgwa2
not modelled
5.6
14
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Biotin holoenzyme synthetase49 d1gzga_
not modelled
5.6
25
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)50 c2kl8A_
not modelled
5.5
50
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
51 d1l0nk_
not modelled
5.5
50
Fold: Single transmembrane helixSuperfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)52 c3kewA_
not modelled
5.5
27
PDB header: transferaseChain: A: PDB Molecule: dhha1 domain protein;PDBTitle: crystal structure of probable alanyl-trna-synthase from clostridium2 perfringens
53 d2e1ba1
not modelled
5.4
47
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: AlaX-M N-terminal domain-like54 d1qy9a1
not modelled
5.3
14
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like55 c3kwmC_
not modelled
5.2
24
PDB header: isomeraseChain: C: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-isomerase a
56 c2kyrA_
not modelled
5.2
19
PDB header: transferaseChain: A: PDB Molecule: fructose-like phosphotransferase enzyme iib component 1;PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
57 d2ifaa1
not modelled
5.1
10
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase58 c3mp2A_
not modelled
5.0
13
PDB header: hydrolaseChain: A: PDB Molecule: non-structural protein 3;PDBTitle: crystal structure of transmissible gastroenteritis virus papain-like2 protease 1