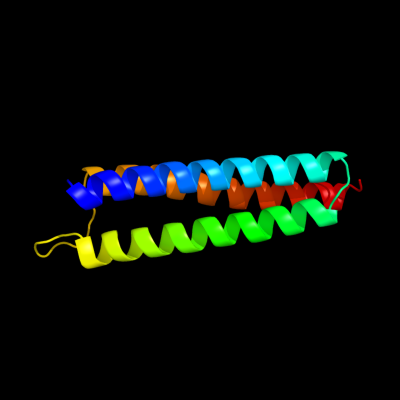
1 c1wcrA_
100.0
99
PDB header: transferaseChain: A: PDB Molecule: pts system, n, n'-diacetylchitobiose-specificPDBTitle: trimeric structure of the enzyme iia from escherichia coli2 phosphotransferase system specific for n,n'-3 diacetylchitobiose
2 d2e2aa_
100.0
33
Fold: Spectrin repeat-likeSuperfamily: Enzyme IIa from lactose specific PTS, IIa-lacFamily: Enzyme IIa from lactose specific PTS, IIa-lac3 c3l8rA_
100.0
35
PDB header: transferaseChain: A: PDB Molecule: putative pts system, cellobiose-specific iiaPDBTitle: the crystal structure of ptca from s. mutans
4 c3k1sE_
100.0
45
PDB header: transferaseChain: E: PDB Molecule: pts system, cellobiose-specific iia component;PDBTitle: crystal structure of the pts cellobiose specific enzyme iia2 from bacillus anthracis
5 c2w2uA_
60.5
26
PDB header: hydrolase/transportChain: A: PDB Molecule: hypothetical p60 katanin;PDBTitle: structural insight into the interaction between archaeal2 escrt-iii and aaa-atpase
6 d2jq9a1
50.2
17
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain7 d1wr0a1
49.1
20
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain8 d2cpta1
46.2
21
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain9 c2v6xA_
43.1
12
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: stractural insight into the interaction between escrt-iii2 and vps4
10 c2v6yA_
43.0
26
PDB header: hydrolaseChain: A: PDB Molecule: aaa family atpase, p60 katanin;PDBTitle: structure of the mit domain from a s. solfataricus vps4-2 like atpase
11 d1wfda_
39.4
11
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain12 c3zxpC_
34.1
13
PDB header: protein transportChain: C: PDB Molecule: bro1 domain-containing protein brox;PDBTitle: structural and functional analyses of the bro1 domain protein brox
13 d2hr2a1
32.2
20
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: CT2138-like14 c3bu8B_
26.9
16
PDB header: dna binding proteinChain: B: PDB Molecule: telomeric repeat-binding factor 2;PDBTitle: crystal structure of trf2 trfh domain and tin2 peptide2 complex
15 d2crba1
26.6
21
Fold: Spectrin repeat-likeSuperfamily: MIT domain-likeFamily: MIT domain16 d1hh8a_
26.3
21
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)17 c2xevB_
24.6
22
PDB header: metal bindingChain: B: PDB Molecule: ybgf;PDBTitle: crystal structure of the tpr domain of xanthomonas2 campestris ybgf
18 d1hxia_
21.8
14
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)19 c1xofA_
21.2
25
PDB header: de novo proteinChain: A: PDB Molecule: bbahett1;PDBTitle: heterooligomeric beta beta alpha miniprotein
20 d2gy9t1
20.3
18
Fold: Spectrin repeat-likeSuperfamily: Ribosomal protein S20Family: Ribosomal protein S2021 d2c2la1
not modelled
17.7
17
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)22 c3r9mA_
not modelled
17.5
13
PDB header: protein bindingChain: A: PDB Molecule: bro1 domain-containing protein brox;PDBTitle: crystal structure of the brox bro1 domain
23 c2avpA_
not modelled
17.2
22
PDB header: de novo proteinChain: A: PDB Molecule: synthetic consensus tpr protein;PDBTitle: crystal structure of an 8 repeat consensus tpr superhelix
24 d2uubt1
not modelled
17.1
21
Fold: Spectrin repeat-likeSuperfamily: Ribosomal protein S20Family: Ribosomal protein S2025 d1elwa_
not modelled
16.1
18
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)26 c2xcbA_
not modelled
14.1
14
PDB header: protein bindingChain: A: PDB Molecule: regulatory protein pcrh;PDBTitle: crystal structure of pcrh in complex with the chaperone2 binding region of popd
27 c2hyzA_
not modelled
14.0
22
PDB header: de novo proteinChain: A: PDB Molecule: synthetic consensus tpr protein;PDBTitle: crystal structure of an 8 repeat consensus tpr superhelix2 (orthorombic crystal form)
28 c1na3A_
not modelled
13.7
21
PDB header: de novo proteinChain: A: PDB Molecule: designed protein ctpr2;PDBTitle: design of stable alpha-helical arrays from an idealized tpr2 motif
29 d2buga1
not modelled
13.4
26
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)30 c2j7aE_
not modelled
13.2
20
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c nitrite reductase nrfa;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
31 c1sfcJ_
not modelled
12.6
12
PDB header: transport proteinChain: J: PDB Molecule: protein (syntaxin 1a);PDBTitle: neuronal synaptic fusion complex
32 d1qhla_
not modelled
12.4
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like33 c2ae3A_
not modelled
12.2
11
PDB header: hydrolaseChain: A: PDB Molecule: glutaryl 7-aminocephalosporanic acid acylase;PDBTitle: glutaryl 7-aminocephalosporanic acid acylase: mutational study of2 activation mechanism
34 c2c2lD_
not modelled
12.1
17
PDB header: chaperoneChain: D: PDB Molecule: carboxy terminus of hsp70-interacting protein;PDBTitle: crystal structure of the chip u-box e3 ubiquitin ligase
35 c2q00B_
not modelled
12.0
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: orf c02003 protein;PDBTitle: crystal structure of the p95883_sulso protein from2 sulfolobus solfataricus. nesg target ssr10.
36 d1tjca_
not modelled
11.9
14
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)37 c3dzaB_
not modelled
11.8
13
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized putative membrane protein;PDBTitle: crystal structure of a putative membrane protein of unknown function2 (yfdx) from klebsiella pneumoniae subsp. at 1.65 a resolution
38 d1wiva_
not modelled
11.5
23
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain39 c2vyiA_
not modelled
11.4
26
PDB header: chaperoneChain: A: PDB Molecule: sgta protein;PDBTitle: crystal structure of the tpr domain of human sgt
40 c3sz7A_
not modelled
11.1
11
PDB header: chaperone regulatorChain: A: PDB Molecule: hsc70 cochaperone (sgt);PDBTitle: crystal structure of the sgt2 tpr domain from aspergillus fumigatus
41 d1ndba2
not modelled
10.9
17
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase42 c3ipdB_
not modelled
10.6
15
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
43 c1gk9A_
not modelled
10.6
37
PDB header: antibiotic resistanceChain: A: PDB Molecule: penicillin g acylase alpha subunit;PDBTitle: crystal structures of penicillin acylase enzyme-substrate2 complexes: structural insights into the catalytic mechanism
44 d2pqrb1
not modelled
10.4
29
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)45 c3ct5A_
not modelled
10.2
15
PDB header: hydrolaseChain: A: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall degrading enzyme2 in the bacteriophage phi29 tail
46 c3oxgA_
not modelled
9.9
19
PDB header: transferaseChain: A: PDB Molecule: set and mynd domain-containing protein 3;PDBTitle: human lysine methyltransferase smyd3 in complex with adohcy (form iii)
47 c3q49B_
not modelled
9.6
14
PDB header: ligase/chaperoneChain: B: PDB Molecule: stip1 homology and u box-containing protein 1;PDBTitle: crystal structure of the tpr domain of chip complexed with hsp70-c2 peptide
48 c1ajnA_
not modelled
9.3
37
PDB header: antibiotic resistanceChain: A: PDB Molecule: penicillin amidohydrolase;PDBTitle: penicillin acylase complexed with p-nitrophenylacetic acid
49 c2npsB_
not modelled
9.3
12
PDB header: transport proteinChain: B: PDB Molecule: syntaxin 13;PDBTitle: crystal structure of the early endosomal snare complex
50 d1xl7a2
not modelled
9.2
15
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase51 c1cp9A_
not modelled
8.7
26
PDB header: hydrolaseChain: A: PDB Molecule: penicillin amidohydrolase;PDBTitle: crystal structure of penicillin g acylase from the bro12 mutant strain of providencia rettgeri
52 c3k3wA_
not modelled
8.6
32
PDB header: hydrolaseChain: A: PDB Molecule: penicillin g acylase;PDBTitle: thermostable penicillin g acylase from alcaligence faecalis2 in orthorhombic form
53 d1a17a_
not modelled
8.5
26
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)54 c2kckA_
not modelled
8.3
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tpr repeat;PDBTitle: nmr solution structure of the northeast structural genomics2 consortium (nesg) target mrr121a
55 d2joka1
not modelled
8.2
23
Fold: SopE-like GEF domainSuperfamily: SopE-like GEF domainFamily: SopE-like GEF domain56 c2if4A_
not modelled
8.0
14
PDB header: signaling proteinChain: A: PDB Molecule: atfkbp42;PDBTitle: crystal structure of a multi-domain immunophilin from2 arabidopsis thaliana
57 d1xb2b1
not modelled
7.9
20
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain58 c2vr0A_
not modelled
7.8
20
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase, catalytic subunit nfra;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex bound to the hqno inhibitor
59 c3qkyA_
not modelled
7.4
13
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane assembly lipoprotein yfio;PDBTitle: crystal structure of rhodothermus marinus bamd
60 c3ceqB_
not modelled
7.4
19
PDB header: motor protein, transport proteinChain: B: PDB Molecule: kinesin light chain 2;PDBTitle: the tpr domain of human kinesin light chain 2 (hklc2)
61 d2choa1
not modelled
7.3
7
Fold: Hyaluronidase domain-likeSuperfamily: Hyaluronidase post-catalytic domain-likeFamily: Hyaluronidase post-catalytic domain-like62 c1n7sB_
not modelled
7.2
9
PDB header: transport proteinChain: B: PDB Molecule: syntaxin 1a;PDBTitle: high resolution structure of a truncated neuronal snare2 complex
63 d2bcgg2
not modelled
7.1
0
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: GDI-like N domain64 c2r5sB_
not modelled
7.0
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp0806;PDBTitle: the crystal structure of a domain of protein vp0806 (unknown function)2 from vibrio parahaemolyticus rimd 2210633
65 c3b5nF_
not modelled
7.0
13
PDB header: membrane proteinChain: F: PDB Molecule: protein sso1;PDBTitle: structure of the yeast plasma membrane snare complex
66 d1aipc1
not modelled
6.9
22
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain67 c2wybA_
not modelled
6.9
11
PDB header: hydrolaseChain: A: PDB Molecule: acyl-homoserine lactone acylase pvdq subunitPDBTitle: the quorum quenching n-acyl homoserine lactone acylase pvdq2 with a covalently bound dodecanoic acid
68 d2cbia1
not modelled
6.6
14
Fold: Hyaluronidase domain-likeSuperfamily: Hyaluronidase post-catalytic domain-likeFamily: Hyaluronidase post-catalytic domain-like69 d1p5qa1
not modelled
6.4
14
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)70 c1kt0A_
not modelled
6.2
17
PDB header: isomeraseChain: A: PDB Molecule: 51 kda fk506-binding protein;PDBTitle: structure of the large fkbp-like protein, fkbp51, involved in steroid2 receptor complexes
71 c2r0bA_
not modelled
6.1
24
PDB header: hydrolaseChain: A: PDB Molecule: serine/threonine/tyrosine-interacting protein;PDBTitle: crystal structure of human tyrosine phosphatase-like2 serine/threonine/tyrosine-interacting protein
72 c3rauB_
not modelled
6.1
21
PDB header: hydrolaseChain: B: PDB Molecule: tyrosine-protein phosphatase non-receptor type 23;PDBTitle: crystal structure of the hd-ptp bro1 domain
73 d1vh6a_
not modelled
5.9
9
Fold: Four-helical up-and-down bundleSuperfamily: Flagellar export chaperone FliSFamily: Flagellar export chaperone FliS74 c1vh6A_
not modelled
5.9
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flagellar protein flis;PDBTitle: crystal structure of a flagellar protein
75 d1iqpa1
not modelled
5.7
17
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain76 d2ivxa2
not modelled
5.7
37
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin77 c1ii8A_
not modelled
5.6
17
PDB header: replicationChain: A: PDB Molecule: rad50 abc-atpase;PDBTitle: crystal structure of the p. furiosus rad50 atpase domain
78 c1efuB_
not modelled
5.6
30
PDB header: complex (two elongation factors)Chain: B: PDB Molecule: elongation factor ts;PDBTitle: elongation factor complex ef-tu/ef-ts from escherichia coli
79 d3blhb1
not modelled
5.6
42
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin80 d256ba_
not modelled
5.5
29
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome b56281 c3kvhA_
not modelled
5.4
14
PDB header: rna binding proteinChain: A: PDB Molecule: protein syndesmos;PDBTitle: crystal structure of human protein syndesmos (nudt16-like protein)
82 d2cp9a1
not modelled
5.4
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain83 c1xexA_
not modelled
5.4
21
PDB header: cell cycleChain: A: PDB Molecule: smc protein;PDBTitle: structural biochemistry of atp-driven dimerization and dna2 stimulated activation of smc atpases.
84 c1ycsB_
not modelled
5.3
16
PDB header: complex (anti-oncogene/ankyrin repeats)Chain: B: PDB Molecule: 53bp2;PDBTitle: p53-53bp2 complex
85 d2idga1
not modelled
5.2
10
Fold: TorD-likeSuperfamily: TorD-likeFamily: TorD-like86 c2oudA_
not modelled
5.2
16
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of the catalytic domain of human mkp5
87 c2kc7A_
not modelled
5.2
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bfr218_protein;PDBTitle: solution nmr structure of bacteroides fragilis protein2 bf1650. northeast structural genomics consortium target3 bfr218
88 c3gyzB_
not modelled
5.1
14
PDB header: chaperoneChain: B: PDB Molecule: chaperone protein ipgc;PDBTitle: crystal structure of ipgc from shigella flexneri