1 d2j3wa1
100.0
100
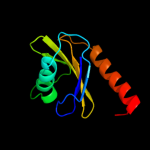
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: Sedlin (SEDL)2 c2j3tC_
100.0
11
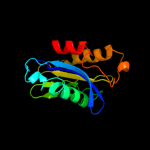
PDB header: transportChain: C: PDB Molecule: trafficking protein particle complex subunit 1;PDBTitle: the crystal structure of the bet3-trs33-bet5-trs23 complex.
3 c3cueM_
100.0
18
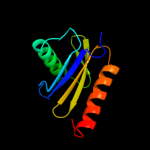
PDB header: protein transportChain: M: PDB Molecule: transport protein particle 23 kda subunit;PDBTitle: crystal structure of a trapp subassembly activating the rab ypt1p
4 c3pr6A_
100.0
17
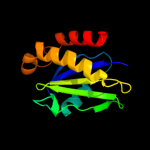
PDB header: transport proteinChain: A: PDB Molecule: trapp-associated protein tca17;PDBTitle: crystal structure analysis of yeast trapp associate protein tca17
5 c2zmvB_
100.0
26
PDB header: transport proteinChain: B: PDB Molecule: trafficking protein particle complex subunit 4;PDBTitle: crystal structure of synbindin
6 c3cueO_
100.0
19
PDB header: protein transportChain: O: PDB Molecule: transport protein particle 18 kda subunit;PDBTitle: crystal structure of a trapp subassembly activating the rab ypt1p
7 c3kyqA_
94.3
8
PDB header: transferaseChain: A: PDB Molecule: synaptobrevin homolog ykt6;PDBTitle: lipid-induced conformational switch controls fusion activity of longin2 domain snare ykt6
8 d1gw5m2
91.2
13
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: Clathrin coat assembly domain9 d1ioua_
86.6
7
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: Synatpobrevin N-terminal domain10 c2hf6A_
84.3
6
PDB header: protein transportChain: A: PDB Molecule: coatomer subunit zeta-1;PDBTitle: solution structure of human zeta-cop
11 c2dmwA_
63.7
13
PDB header: membrane proteinChain: A: PDB Molecule: synaptobrevin-like 1 variant;PDBTitle: solution structure of the longin domain of synaptobrevin-2 like protein 1
12 c2jkrM_
63.4
13
PDB header: endocytosisChain: M: PDB Molecule: ap-2 complex subunit mu-1;PDBTitle: ap2 clathrin adaptor core with dileucine peptide rm(2 phosphos)qikrllse
13 c3egdC_
61.7
9
PDB header: protein transportChain: C: PDB Molecule: vesicle-trafficking protein sec22b;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23a/24a complexed with the snare protein sec22 and bound3 to the transport signal sequence of vesicular stomatitis4 virus glycoprotein
14 c2nupC_
61.7
9
PDB header: protein transportChain: C: PDB Molecule: vesicle-trafficking protein sec22b;PDBTitle: crystal structure of the human sec23a/24a heterodimer,2 complexed with the snare protein sec22b
15 c1w63P_
47.0
3
PDB header: endocytosisChain: P: PDB Molecule: adaptor-related protein complex 1, mu 1 subunit;PDBTitle: ap1 clathrin adaptor core
16 c4b93A_
42.7
13
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 7;PDBTitle: complex of vamp7 cytoplasmic domain with 2nd ankyrin repeat2 domain of varp
17 d1ifqa_
40.3
10
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: Synatpobrevin N-terminal domain18 c1mp9B_
21.0
11
PDB header: dna binding proteinChain: B: PDB Molecule: tata-binding protein;PDBTitle: tbp from a mesothermophilic archaeon, sulfolobus2 acidocaldarius
19 d1gw5s_
20.4
11
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: Clathrin coat assembly domain20 c4b0aA_
18.0
14
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation factor tfiid subunit 1, linker,PDBTitle: the high-resolution structure of ytbp-ytaf1 identifies2 conserved and competing interaction surfaces in3 transcriptional activation
21 c1d3uA_
not modelled
17.7
8
PDB header: gene regulation/dnaChain: A: PDB Molecule: tata-binding protein;PDBTitle: tata-binding protein/transcription factor (ii)b/bre+tata-2 box complex from pyrococcus woesei
22 c4eiyA_
not modelled
17.5
23
PDB header: membrane proteinChain: A: PDB Molecule: adenosine receptor a2a/soluble cytochrome b562 chimera;PDBTitle: crystal structure of the chimeric protein of a2aar-bril in complex2 with zm241385 at 1.8a resolution
23 c1yd6A_
not modelled
17.4
13
PDB header: dna binding proteinChain: A: PDB Molecule: uvrc;PDBTitle: crystal structure of the giy-yig n-terminal endonuclease2 domain of uvrc from bacillus caldotenax
24 c1rm1A_
not modelled
17.1
14
PDB header: transcription/dnaChain: A: PDB Molecule: tata-box binding protein;PDBTitle: structure of a yeast tfiia/tbp/tata-box dna complex
25 c1ngmM_
not modelled
17.1
14
PDB header: transcription/dnaChain: M: PDB Molecule: transcription initiation factor tfiid;PDBTitle: crystal structure of a yeast brf1-tbp-dna ternary complex
26 c1w63T_
not modelled
15.2
6
PDB header: endocytosisChain: T: PDB Molecule: adapter-related protein complex 1 sigma 1aPDBTitle: ap1 clathrin adaptor core
27 c2z8uQ_
not modelled
13.6
10
PDB header: transcriptionChain: Q: PDB Molecule: tata-box-binding protein;PDBTitle: methanococcus jannaschii tbp
28 d1e0na_
not modelled
13.6
17
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain29 c3eikB_
not modelled
12.9
14
PDB header: transcriptionChain: B: PDB Molecule: tata-box-binding protein;PDBTitle: double stranded dna binding protein
30 c3trjC_
not modelled
11.5
10
PDB header: isomeraseChain: C: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
31 c3bcyA_
not modelled
10.9
39
PDB header: unknown functionChain: A: PDB Molecule: protein yer067w;PDBTitle: crystal structure of yer067w
32 c3emlA_
not modelled
10.6
24
PDB header: membrane protein, receptorChain: A: PDB Molecule: human adenosine a2a receptor/t4 lysozyme chimera;PDBTitle: the 2.6 a crystal structure of a human a2a adenosine receptor bound to2 zm241385.
33 c4ax2A_
not modelled
9.5
27
PDB header: toxinChain: A: PDB Molecule: rap1b;PDBTitle: new type vi-secreted toxins and self-resistance proteins in2 serratia marcescens
34 d1qnaa2
not modelled
9.2
10
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain35 d2cxha1
not modelled
9.0
22
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Brix domain36 c2cxhA_
not modelled
9.0
22
PDB header: rna binding proteinChain: A: PDB Molecule: probable brix-domain ribosomal biogenesis protein;PDBTitle: crystal structure of probable ribosomal biogenesis protein from2 aeropyrum pernix k1
37 d1cdwa2
not modelled
7.4
10
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain38 d1es6a1
not modelled
7.1
29
Fold: EV matrix proteinSuperfamily: EV matrix proteinFamily: EV matrix protein39 c3tcqA_
not modelled
6.9
29
PDB header: viral proteinChain: A: PDB Molecule: matrix protein vp40;PDBTitle: crystal structure of matrix protein vp40 from ebola virus sudan
40 d1h2ca_
not modelled
6.9
29
Fold: EV matrix proteinSuperfamily: EV matrix proteinFamily: EV matrix protein41 c3rzeA_
not modelled
6.8
30
PDB header: hydrolaseChain: A: PDB Molecule: histamine h1 receptor, lysozyme chimera;PDBTitle: structure of the human histamine h1 receptor in complex with doxepin
42 c1h2dA_
not modelled
6.7
29
PDB header: virus/viral proteinChain: A: PDB Molecule: matrix protein vp40;PDBTitle: ebola virus matrix protein vp40 n-terminal domain in2 complex with rna (low-resolution vp40[31-212] variant).
43 c2go51_
not modelled
6.5
10
PDB header: translation/rnaChain: 1: PDB Molecule: signal recognition particle receptor alphaPDBTitle: structure of signal recognition particle receptor (sr) in2 complex with signal recognition particle (srp) and3 ribosome nascent chain complex
44 d1d2zb_
not modelled
5.6
16
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD45 c1z5sD_
not modelled
5.1
23
PDB header: ligaseChain: D: PDB Molecule: ran-binding protein 2;PDBTitle: crystal structure of a complex between ubc9, sumo-1,2 rangap1 and nup358/ranbp2