| 1 | d2g9da1
|
|
 |
100.0 |
34 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 2 | d1yw6a1
|
|
 |
100.0 |
97 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 3 | d2bcoa1
|
|
 |
100.0 |
32 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 4 | d1yw4a1
|
|
 |
100.0 |
38 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 5 | d2gu2a1
|
|
 |
100.0 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 6 | c3nh8A_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:aspartoacylase-2;
PDBTitle: crystal structure of murine aminoacylase 3 in complex with n-acetyl-s-2 1,2-dichlorovinyl-l-cysteine
|
| 7 | d2i3ca1
|
|
 |
100.0 |
18 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 8 | c3na6A_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:succinylglutamate desuccinylase/aspartoacylase;
PDBTitle: crystal structure of a succinylglutamate desuccinylase (tm1040_2694)2 from silicibacter sp. tm1040 at 2.00 a resolution
|
| 9 | c3cdxB_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:succinylglutamatedesuccinylase/aspartoacylase;
PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
|
| 10 | c3fmcC_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative succinylglutamate desuccinylase / aspartoacylase;
PDBTitle: crystal structure of a putative succinylglutamate desuccinylase /2 aspartoacylase family protein (sama_0604) from shewanella amazonensis3 sb2b at 1.80 a resolution
|
| 11 | c2qj8B_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:mlr6093 protein;
PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
|
| 12 | c2qvpC_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative metallopeptidase (sama_0725) from2 shewanella amazonensis sb2b at 2.00 a resolution
|
| 13 | c3b2yB_
|
|
 |
99.9 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:metallopeptidase containing co-catalytic metalloactive
PDBTitle: crystal structure of a putative metallopeptidase (sden_2526) from2 shewanella denitrificans os217 at 1.74 a resolution
|
| 14 | c3d4uA_
|
|
 |
99.5 |
17 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:carboxypeptidase b2;
PDBTitle: bovine thrombin-activatable fibrinolysis inhibitor (tafia) in complex2 with tick-derived carboxypeptidase inhibitor.
|
| 15 | d1z5ra1
|
|
 |
99.5 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 16 | d1kwma1
|
|
 |
99.5 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 17 | d1nsaa1
|
|
 |
99.5 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 18 | d1obra_
|
|
 |
99.5 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Carboxypeptidase T |
| 19 | c3mn8A_
|
|
 |
99.4 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:lp15968p;
PDBTitle: structure of drosophila melanogaster carboxypeptidase d isoform 1b2 short
|
| 20 | d1jqga1
|
|
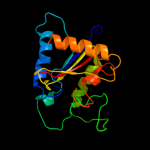 |
99.4 |
21 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 21 | d1h8la2 |
|
not modelled |
99.4 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 22 | d1m4la_ |
|
not modelled |
99.4 |
21 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 23 | d2bo9a1 |
|
not modelled |
99.4 |
26 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 24 | c3l2nA_ |
|
not modelled |
99.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase m14, carboxypeptidase a;
PDBTitle: crystal structure of putative carboxypeptidase a (yp_562911.1) from2 shewanella denitrificans os-217 at 2.39 a resolution
|
| 25 | c1nsaA_ |
|
not modelled |
99.4 |
20 |
PDB header:serine protease
Chain: A: PDB Molecule:procarboxypeptidase b;
PDBTitle: three-dimensional structure of porcine procarboxypeptidase2 b: a structural basis of its inactivity
|
| 26 | d1ayea1 |
|
not modelled |
99.4 |
21 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 27 | c1h8lA_ |
|
not modelled |
99.4 |
18 |
PDB header:carboxypeptidase
Chain: A: PDB Molecule:carboxypeptidase gp180 residues 503-882;
PDBTitle: duck carboxypeptidase d domain ii in complex with gemsa
|
| 28 | d2c1ca1 |
|
not modelled |
99.4 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 29 | c1uwyA_ |
|
not modelled |
99.3 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxypeptidase m;
PDBTitle: crystal structure of human carboxypeptidase m
|
| 30 | d1uwya2 |
|
not modelled |
99.3 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Pancreatic carboxypeptidases |
| 31 | c1jqgA_ |
|
not modelled |
99.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxypeptidase a;
PDBTitle: crystal structure of the carboxypeptidase a from2 helicoverpa armigera
|
| 32 | c2boaB_ |
|
not modelled |
99.3 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:carboxypeptidase a4;
PDBTitle: human procarboxypeptidase a4.
|
| 33 | c3k2kA_ |
|
not modelled |
99.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative carboxypeptidase;
PDBTitle: crystal structure of putative carboxypeptidase (yp_103406.1) from2 burkholderia mallei atcc 23344 at 2.49 a resolution
|
| 34 | c1ayeA_ |
|
not modelled |
99.3 |
21 |
PDB header:serine protease
Chain: A: PDB Molecule:procarboxypeptidase a2;
PDBTitle: human procarboxypeptidase a2
|
| 35 | c3dgvB_ |
|
not modelled |
99.2 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:carboxypeptidase b2;
PDBTitle: crystal structure of thrombin activatable fibrinolysis inhibitor2 (tafi)
|
| 36 | c2nsmA_ |
|
not modelled |
99.2 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxypeptidase n catalytic chain;
PDBTitle: crystal structure of the human carboxypeptidase n (kininase i)2 catalytic domain
|
| 37 | d2q7sa1 |
|
not modelled |
96.3 |
15 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:FGase-like |
| 38 | c1cpbB_ |
|
not modelled |
96.0 |
21 |
PDB header:hydrolase (c-terminal peptidase)
Chain: B: PDB Molecule:carboxypeptidase b;
PDBTitle: structure of carboxypeptidase b at 2.8 angstroms resolution
|
| 39 | c1cpbA_ |
|
not modelled |
95.5 |
12 |
PDB header:hydrolase (c-terminal peptidase)
Chain: A: PDB Molecule:carboxypeptidase b;
PDBTitle: structure of carboxypeptidase b at 2.8 angstroms resolution
|
| 40 | d2odfa1 |
|
not modelled |
91.8 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:FGase-like |
| 41 | c1m6bA_ |
|
not modelled |
43.1 |
9 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:receptor protein-tyrosine kinase erbb-3;
PDBTitle: structure of the her3 (erbb3) extracellular domain
|
| 42 | c1nqlA_ |
|
not modelled |
40.7 |
11 |
PDB header:hormone/growth factor receptor
Chain: A: PDB Molecule:epidermal growth factor receptor;
PDBTitle: structure of the extracellular domain of human epidermal growth factor2 (egf) receptor in an inactive (low ph) complex with egf.
|
| 43 | c3qayC_ |
|
not modelled |
38.3 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:endolysin;
PDBTitle: catalytic domain of cd27l endolysin targeting clostridia difficile
|
| 44 | c3ltgA_ |
|
not modelled |
38.3 |
19 |
PDB header:transferase/transferase regulator
Chain: A: PDB Molecule:epidermal growth factor receptor;
PDBTitle: crystal structure of the drosophila epidermal growth factor receptor2 ectodomain complexed with a low affinity spitz mutant
|
| 45 | c3be1A_ |
|
not modelled |
31.6 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: dual specific bh1 fab in complex with the extracellular domain of2 her2/erbb-2
|
| 46 | c3i2tA_ |
|
not modelled |
30.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:epidermal growth factor receptor, isoform a;
PDBTitle: crystal structure of the unliganded drosophila epidermal growth factor2 receptor ectodomain
|
| 47 | d1jwqa_ |
|
not modelled |
26.6 |
14 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 48 | c3p11A_ |
|
not modelled |
21.5 |
11 |
PDB header:immune system
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: anti-egfr/her3 fab dl11 in complex with domains i-iii of the her32 extracellular region
|
| 49 | c2q5cA_ |
|
not modelled |
19.2 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 50 | c1bdeA_ |
|
not modelled |
18.1 |
26 |
PDB header:aids
Chain: A: PDB Molecule:vpr protein;
PDBTitle: helical structure of polypeptides from the c-terminal half2 of hiv-1 vpr, nmr, 20 structures
|
| 51 | c1xovA_ |
|
not modelled |
18.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:ply protein;
PDBTitle: the crystal structure of the listeria monocytogenes bacteriophage psa2 endolysin plypsa
|
| 52 | d1xova2 |
|
not modelled |
17.7 |
15 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:N-acetylmuramoyl-L-alanine amidase-like |
| 53 | c2ahxB_ |
|
not modelled |
17.2 |
8 |
PDB header:cell cycle,signaling protein
Chain: B: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: crystal structure of erbb4/her4 extracellular domain
|
| 54 | c2hklB_ |
|
not modelled |
15.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:l,d-transpeptidase;
PDBTitle: crystal structure of enterococcus faecium l,d-2 transpeptidase c442s mutant
|
| 55 | c3czxA_ |
|
not modelled |
14.6 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative n-acetylmuramoyl-l-alanine amidase;
PDBTitle: the crystal structure of the putative n-acetylmuramoyl-l-2 alanine amidase from neisseria meningitidis
|
| 56 | c3lkxB_ |
|
not modelled |
13.3 |
13 |
PDB header:chaperone
Chain: B: PDB Molecule:nascent polypeptide-associated complex subunit alpha;
PDBTitle: human nac dimerization domain
|
| 57 | d1no7a_ |
|
not modelled |
10.9 |
23 |
Fold:Major capsid protein VP5
Superfamily:Major capsid protein VP5
Family:Major capsid protein VP5 |
| 58 | c1no7A_ |
|
not modelled |
10.9 |
23 |
PDB header:viral protein
Chain: A: PDB Molecule:major capsid protein;
PDBTitle: structure of the large protease resistant upper domain of2 vp5, the major capsid protein of herpes simplex virus-1
|
| 59 | c1dsjA_ |
|
not modelled |
10.4 |
28 |
PDB header:viral peptide
Chain: A: PDB Molecule:vpr protein;
PDBTitle: nmr solution structure of vpr50_75, 20 structures
|
| 60 | d2ix0a3 |
|
not modelled |
8.0 |
26 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 61 | d2ihoa2 |
|
not modelled |
7.6 |
42 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:MOA C-terminal domain-like |
| 62 | c1moxB_ |
|
not modelled |
7.2 |
11 |
PDB header:transferase/growth factor
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: crystal structure of human epidermal growth factor receptor (residues2 1-501) in complex with tgf-alpha
|
| 63 | d1g5ba_ |
|
not modelled |
6.5 |
22 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 64 | c1igrA_ |
|
not modelled |
6.5 |
28 |
PDB header:hormone receptor
Chain: A: PDB Molecule:insulin-like growth factor receptor 1;
PDBTitle: type 1 insulin-like growth factor receptor (domains 1-3)
|
| 65 | d2grea2 |
|
not modelled |
6.1 |
11 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:Bacterial dinuclear zinc exopeptidases |
| 66 | c3pctA_ |
|
not modelled |
5.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:class c acid phosphatase;
PDBTitle: structure of the class c acid phosphatase from pasteurella multocida
|
| 67 | c3et4A_ |
|
not modelled |
5.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:outer membrane protein p4, nadp phosphatase;
PDBTitle: structure of recombinant haemophilus influenzae e(p4) acid phosphatase
|