| 1 | d2csga1 |
|
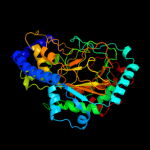 |
100.0 |
85 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:YbiU-like |
| 2 | c2dbiA_ |
|
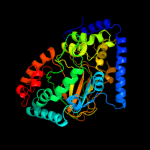 |
100.0 |
98 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ybiu;
PDBTitle: crystal structure of a hypothetical protein jw0805 from2 escherichia coli
|
| 3 | c2opwA_ |
|
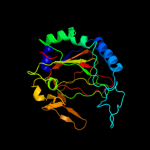 |
99.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phyhd1 protein;
PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
|
| 4 | d2fcta1 |
|
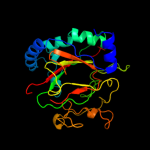 |
99.2 |
15 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 5 | d2a1xa1 |
|
 |
99.2 |
16 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 6 | c3gjbA_ |
|
 |
99.1 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cytc3;
PDBTitle: cytc3 with fe(ii) and alpha-ketoglutarate
|
| 7 | c3emrA_ |
|
 |
99.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ectd;
PDBTitle: crystal structure analysis of the ectoine hydroxylase ectd from2 salibacillus salexigens
|
| 8 | c2rdsA_ |
|
 |
98.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxypentalenic acid 11-beta hydroxylase; fe(ii)/alpha-
PDBTitle: crystal structure of ptlh with fe/oxalylglycine and ent-1-2 deoxypentalenic acid bound
|
| 9 | c3nnlB_ |
|
 |
98.0 |
18 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cura;
PDBTitle: halogenase domain from cura module (crystal form iii)
|
| 10 | d1dcsa_ |
|
 |
96.7 |
16 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 11 | c3on7C_ |
|
 |
95.9 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, iron/ascorbate family;
PDBTitle: crystal structure of a putative oxygenase (so_2589) from shewanella2 oneidensis at 2.20 a resolution
|
| 12 | c3ooxA_ |
|
 |
95.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 2og-fe(ii) oxygenase family protein;
PDBTitle: crystal structure of a putative 2og-fe(ii) oxygenase family protein2 (cc_0200) from caulobacter crescentus at 1.44 a resolution
|
| 13 | d1w9ya1 |
|
 |
90.1 |
13 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 14 | c3dkqB_ |
|
 |
89.8 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pkhd-type hydroxylase sbal_3634;
PDBTitle: crystal structure of putative oxygenase (yp_001051978.1) from2 shewanella baltica os155 at 2.26 a resolution
|
| 15 | c3e77A_ |
|
 |
89.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: human phosphoserine aminotransferase in complex with plp
|
| 16 | c2g19A_ |
|
 |
87.4 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:egl nine homolog 1;
PDBTitle: cellular oxygen sensing: crystal structure of hypoxia-2 inducible factor prolyl hydroxylase (phd2)
|
| 17 | c3ms5A_ |
|
 |
78.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gamma-butyrobetaine dioxygenase;
PDBTitle: crystal structure of human gamma-butyrobetaine,2-oxoglutarate2 dioxygenase 1 (bbox1)
|
| 18 | d1gp6a_ |
|
 |
77.5 |
11 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 19 | c3m5uA_ |
|
 |
76.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: crystal structure of phosphoserine aminotransferase from2 campylobacter jejuni
|
| 20 | d2c0ra1 |
|
 |
76.1 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 21 | c3qm2A_ |
|
not modelled |
68.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
|
| 22 | d1odma_ |
|
not modelled |
66.3 |
7 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Penicillin synthase-like |
| 23 | d1w23a_ |
|
not modelled |
65.0 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 24 | d2z1ca1 |
|
not modelled |
57.1 |
25 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 25 | c2jijA_ |
|
not modelled |
51.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:prolyl-4 hydroxylase;
PDBTitle: crystal structure of the apo form of chlamydomonas2 reinhardtii prolyl-4 hydroxylase type i
|
| 26 | d1v70a_ |
|
not modelled |
49.7 |
18 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1287-like |
| 27 | c3ouiA_ |
|
not modelled |
49.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:egl nine homolog 1;
PDBTitle: phd2-r717 with 40787422
|
| 28 | c3pvjB_ |
|
not modelled |
48.5 |
3 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of the fe(ii)/alpha-ketoglutarate dependent taurine2 dioxygenase from pseudomonas putida kt2440
|
| 29 | d1y9qa2 |
|
not modelled |
44.9 |
24 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Probable transcriptional regulator VC1968, C-terminal domain |
| 30 | d2toda1 |
|
not modelled |
44.8 |
14 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 31 | c3gt2A_ |
|
not modelled |
44.1 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 32 | d1bjna_ |
|
not modelled |
44.0 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 33 | c3pbiA_ |
|
not modelled |
43.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 34 | d2ot2a1 |
|
not modelled |
43.3 |
26 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 35 | c3itqB_ |
|
not modelled |
43.2 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prolyl 4-hydroxylase, alpha subunit domain protein;
PDBTitle: crystal structure of a prolyl 4-hydroxylase from bacillus anthracis
|
| 36 | d1otja_ |
|
not modelled |
42.8 |
6 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 37 | c2a1dH_ |
|
not modelled |
42.5 |
11 |
PDB header:hydrolase/hydrolase inhibitor
Chain: H: PDB Molecule:staphylocoagulase;
PDBTitle: staphylocoagulase bound to bovine thrombin
|
| 38 | c3r1jB_ |
|
not modelled |
42.1 |
6 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
|
| 39 | d2bnma2 |
|
not modelled |
41.8 |
19 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1459-like |
| 40 | c2doqD_ |
|
not modelled |
40.5 |
43 |
PDB header:cell cycle
Chain: D: PDB Molecule:sfi1p;
PDBTitle: crystal structure of sfi1p/cdc31p complex
|
| 41 | d1bw0a_ |
|
not modelled |
40.3 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 42 | d1q3qa2 |
|
not modelled |
40.2 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
| 43 | d1oiha_ |
|
not modelled |
39.6 |
10 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 44 | c2hbkA_ |
|
not modelled |
37.1 |
12 |
PDB header:hydrolase, gene regulation
Chain: A: PDB Molecule:exosome complex exonuclease rrp6;
PDBTitle: structure of the yeast nuclear exosome component, rrp6p,2 reveals an interplay between the active site and the hrdc3 domain; protein in complex with mn
|
| 45 | d7odca1 |
|
not modelled |
36.9 |
27 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 46 | c3sahA_ |
|
not modelled |
35.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:exosome component 10;
PDBTitle: crystal structure of the human rrp6 catalytic domain with y436a2 mutation in the catalytic site
|
| 47 | c3cewA_ |
|
not modelled |
35.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized cupin protein;
PDBTitle: crystal structure of a cupin protein (bf4112) from bacteroides2 fragilis. northeast structural genomics consortium target bfr205
|
| 48 | c3d3rA_ |
|
not modelled |
35.0 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
| 49 | d1g7oa1 |
|
not modelled |
34.8 |
12 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 50 | c3h7yA_ |
|
not modelled |
34.6 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:bacilysin biosynthesis protein bacb;
PDBTitle: crystal structure of bacb, an enzyme involved in bacilysin synthesis,2 in tetragonal form
|
| 51 | d3d3ra1 |
|
not modelled |
34.2 |
21 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 52 | c2on3A_ |
|
not modelled |
33.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:ornithine decarboxylase;
PDBTitle: a structural insight into the inhibition of human and2 leishmania donovani ornithine decarboxylases by 3-aminooxy-3 1-aminopropane
|
| 53 | c3cp8C_ |
|
not modelled |
33.3 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: crystal structure of gida from chlorobium tepidum
|
| 54 | d1vp4a_ |
|
not modelled |
33.3 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 55 | d2hbka2 |
|
not modelled |
31.6 |
12 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:DnaQ-like 3'-5' exonuclease |
| 56 | c3kt4A_ |
|
not modelled |
30.9 |
5 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pkhd-type hydroxylase tpa1;
PDBTitle: crystal structure of tpa1 from saccharomyces cerevisiae, a2 component of the messenger ribonucleoprotein complex
|
| 57 | d1f3ta1 |
|
not modelled |
30.7 |
14 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 58 | c2xivA_ |
|
not modelled |
30.4 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 59 | d2evra2 |
|
not modelled |
30.1 |
43 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 60 | c3fcrA_ |
|
not modelled |
27.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:putative aminotransferase;
PDBTitle: crystal structure of putative aminotransferase (yp_614685.1) from2 silicibacter sp. tm1040 at 1.80 a resolution
|
| 61 | d1njjb1 |
|
not modelled |
27.7 |
14 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 62 | d1sfna_ |
|
not modelled |
27.6 |
16 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:YlbA-like |
| 63 | d1pmma_ |
|
not modelled |
27.2 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Pyridoxal-dependent decarboxylase |
| 64 | c3lnoA_ |
|
not modelled |
26.8 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
|
| 65 | c3bq9A_ |
|
not modelled |
26.6 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
| 66 | c3dxvA_ |
|
not modelled |
26.4 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:alpha-amino-epsilon-caprolactam racemase;
PDBTitle: the crystal structure of alpha-amino-epsilon-caprolactam racemase from2 achromobacter obae
|
| 67 | c3thtB_ |
|
not modelled |
25.9 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkylated dna repair protein alkb homolog 8;
PDBTitle: crystal structure and rna binding properties of the rrm/alkb domains2 in human abh8, an enzyme catalyzing trna hypermodification, northeast3 structural genomics consortium target hr5601b
|
| 68 | d1uwda_ |
|
not modelled |
25.6 |
14 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:PaaD-like |
| 69 | c2fg0B_ |
|
not modelled |
25.4 |
43 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 70 | c1knwA_ |
|
not modelled |
25.3 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase
|
| 71 | c3ibmB_ |
|
not modelled |
24.4 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:cupin 2, conserved barrel domain protein;
PDBTitle: crystal structure of cupin 2 domain-containing protein hhal_0468 from2 halorhodospira halophila
|
| 72 | c2wl2B_ |
|
not modelled |
24.3 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit a;
PDBTitle: crystal structure of n-terminal domain of gyra with the2 antibiotic simocyclinone d8
|
| 73 | c2ozjB_ |
|
not modelled |
22.8 |
22 |
PDB header:unknown function
Chain: B: PDB Molecule:cupin 2, conserved barrel;
PDBTitle: crystal structure of a cupin superfamily protein (dsy2733) from2 desulfitobacterium hafniense dcb-2 at 1.60 a resolution
|
| 74 | c2xcqA_ |
|
not modelled |
22.8 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:dna gyrase subunit b, dna gyrase subunit a;
PDBTitle: the 2.98a crystal structure of the catalytic core (b'a'2 region) of staphylococcus aureus dna gyrase
|
| 75 | c3s6eB_ |
|
not modelled |
22.5 |
23 |
PDB header:rna binding protein
Chain: B: PDB Molecule:rna-binding protein 39;
PDBTitle: crystal structure of a rna binding motif protein 39 (rbm39) from mus2 musculus at 0.95 a resolution
|
| 76 | c3t18D_ |
|
not modelled |
22.2 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:aminotransferase class i and ii;
PDBTitle: crystal structure of aminotransferase from anaerococcus prevotii dsm2 20548.
|
| 77 | c3h41A_ |
|
not modelled |
22.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60 family protein;
PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
|
| 78 | c3h8uA_ |
|
not modelled |
21.5 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein with double-stranded
PDBTitle: crystal structure of uncharacterized conserved protein with double-2 stranded beta-helix domain (yp_001338853.1) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.80 a resolution
|
| 79 | c3jzvA_ |
|
not modelled |
21.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein rru_a2000;
PDBTitle: crystal structure of rru_a2000 from rhodospirillum rubrum: a cupin-22 domain.
|
| 80 | d1d7ka1 |
|
not modelled |
20.9 |
19 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 81 | d2mev1_ |
|
not modelled |
20.6 |
19 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
| 82 | d1t0fa2 |
|
not modelled |
20.6 |
20 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:TnsA endonuclease, N-terminal domain |
| 83 | c2pfyA_ |
|
not modelled |
20.5 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of dctp7, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
|
| 84 | d1apaa_ |
|
not modelled |
20.3 |
11 |
Fold:Ribosome inactivating proteins (RIP)
Superfamily:Ribosome inactivating proteins (RIP)
Family:Plant cytotoxins |
| 85 | c3dnfB_ |
|
not modelled |
20.3 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
| 86 | c2xcsD_ |
|
not modelled |
20.1 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:dna gyrase subunit b, dna gyrase subunit a;
PDBTitle: the 2.1a crystal structure of s. aureus gyrase complex with gsk2994232 and dna
|
| 87 | c2kp6A_ |
|
not modelled |
19.9 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of protein cv0237 from2 chromobacterium violaceum. northeast structural genomics3 consortium (nesg) target cvt1
|
| 88 | c3dydB_ |
|
not modelled |
19.8 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:tyrosine aminotransferase;
PDBTitle: human tyrosine aminotransferase
|
| 89 | c2pb2B_ |
|
not modelled |
19.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:acetylornithine/succinyldiaminopimelate aminotransferase;
PDBTitle: structure of biosynthetic n-acetylornithine aminotransferase from2 salmonella typhimurium: studies on substrate specificity and3 inhibitor binding
|
| 90 | c2vpvA_ |
|
not modelled |
19.7 |
3 |
PDB header:cell cycle
Chain: A: PDB Molecule:protein mif2;
PDBTitle: dimerization domain of mif2p
|
| 91 | d1knwa1 |
|
not modelled |
19.4 |
22 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 92 | c3fjsC_ |
|
not modelled |
18.8 |
13 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:uncharacterized protein with rmlc-like cupin fold;
PDBTitle: crystal structure of a putative biosynthetic protein with rmlc-like2 cupin fold (reut_b4087) from ralstonia eutropha jmp134 at 1.90 a3 resolution
|
| 93 | d1y0ka1 |
|
not modelled |
18.6 |
41 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:PA4535-like |
| 94 | c3uyjA_ |
|
not modelled |
18.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific demethylase 8;
PDBTitle: crystal structure of jmjd5 catalytic core domain in complex with2 nickle and alpha-kg
|
| 95 | c1e40A_ |
|
not modelled |
18.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: tris/maltotriose complex of chimaeric amylase from b.2 amyloliquefaciens and b. licheniformis at 2.2a
|
| 96 | d1zvpa1 |
|
not modelled |
17.9 |
60 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:VC0802-like |
| 97 | d1bvua2 |
|
not modelled |
17.4 |
18 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 98 | d1gtma2 |
|
not modelled |
16.7 |
18 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 99 | c2p3eA_ |
|
not modelled |
16.4 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of aq1208 from aquifex aeolicus
|

