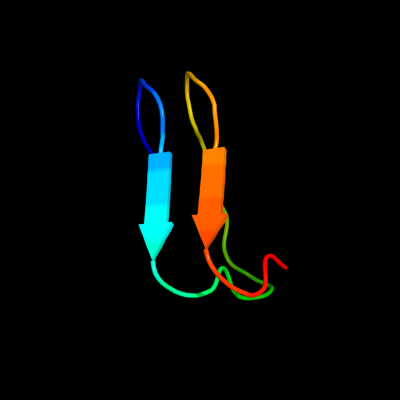
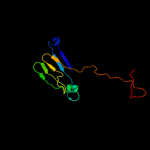
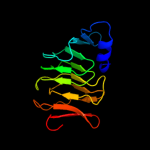
1 d1hf2a1
42.4
32
Fold: Single-stranded right-handed beta-helixSuperfamily: Cell-division inhibitor MinC, C-terminal domainFamily: Cell-division inhibitor MinC, C-terminal domain2 d1jqga1
23.7
20
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Pancreatic carboxypeptidases3 c3st1A_
23.4
21
PDB header: toxinChain: A: PDB Molecule: necrosis-and ethylene-inducing protein;PDBTitle: crystal structure of necrosis and ethylene inducing protein 2 from the2 causal agent of cocoa's witches broom disease
4 c3f6oB_
22.1
8
PDB header: transcription regulatorChain: B: PDB Molecule: probable transcriptional regulator, arsr familyPDBTitle: crystal structure of arsr family transcriptional regulator,2 rha00566
5 d1vjta1
19.3
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like6 c1bdgA_
18.3
20
PDB header: hexokinaseChain: A: PDB Molecule: hexokinase;PDBTitle: hexokinase from schistosoma mansoni complexed with glucose
7 d1vi1a_
17.2
19
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: PlsX-like8 d1w36c1
16.3
42
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain9 c2e72A_
14.2
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pogo transposable element with znf domain;PDBTitle: solution structure of the zinc finger domain of human2 kiaa0461
10 c3u80A_
13.3
23
PDB header: unknown functionChain: A: PDB Molecule: 3-dehydroquinate dehydratase, type ii;PDBTitle: 1.60 angstrom resolution crystal structure of a 3-dehydroquinate2 dehydratase-like protein from bifidobacterium longum
11 c2wycB_
12.9
15
PDB header: hydrolaseChain: B: PDB Molecule: acyl-homoserine lactone acylase pvdq subunitPDBTitle: the quorum quenching n-acyl homoserine lactone acylase pvdq2 in complex with 3-oxo-lauric acid
12 d1ls9a_
12.5
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c13 c3bvsA_
12.2
28
PDB header: hydrolaseChain: A: PDB Molecule: alkylpurine dna glycosylase alkd;PDBTitle: crystal structure of bacillus cereus alkylpurine dna glycosylase alkd
14 d1ef4a_
11.5
42
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB1015 d2q1ma1
10.9
15
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like16 c2ei5B_
10.7
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ttha0061;PDBTitle: crystal structure of hypothetical protein(ttha0061) from thermus2 thermophilus
17 c1hf2A_
10.3
19
PDB header: cell division proteinChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the bacterial cell-division inhibitor2 minc from t. maritima
18 d2k7ia1
10.1
27
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like19 c2k7iB_
10.1
27
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: upf0339 protein atu0232;PDBTitle: solution nmr structure of protein atu0232 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att3. ontario center for structural proteomics target atc0223.
20 d1g3wa2
9.7
17
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain21 c1vjtA_
not modelled
9.0
19
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
22 c3lwzC_
not modelled
8.8
32
PDB header: lyaseChain: C: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
23 c2dxbR_
not modelled
8.7
32
PDB header: hydrolaseChain: R: PDB Molecule: thiocyanate hydrolase subunit gamma;PDBTitle: recombinant thiocyanate hydrolase comprising partially-modified cobalt2 centers
24 c2pmzN_
not modelled
8.5
50
PDB header: translation, transferaseChain: N: PDB Molecule: dna-directed rna polymerase subunit n;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
25 d1uqra_
not modelled
8.4
36
Fold: Flavodoxin-likeSuperfamily: Type II 3-dehydroquinate dehydrataseFamily: Type II 3-dehydroquinate dehydratase26 c3s29C_
not modelled
8.4
22
PDB header: transferaseChain: C: PDB Molecule: sucrose synthase 1;PDBTitle: the crystal structure of sucrose synthase-1 from arabidopsis thaliana2 and its functional implications.
27 d1bu8a1
not modelled
8.2
25
Fold: Lipase/lipooxygenase domain (PLAT/LH2 domain)Superfamily: Lipase/lipooxygenase domain (PLAT/LH2 domain)Family: Colipase-binding domain28 d2fm9a1
not modelled
8.1
47
Fold: SipA N-terminal domain-likeSuperfamily: SipA N-terminal domain-likeFamily: SipA N-terminal domain-like29 d2defa_
not modelled
8.1
19
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase30 d1twfj_
not modelled
8.1
50
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB1031 d2isya2
not modelled
7.9
11
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain32 c3kipU_
not modelled
7.9
32
PDB header: lyaseChain: U: PDB Molecule: 3-dehydroquinase, type ii;PDBTitle: crystal structure of type-ii 3-dehydroquinase from c. albicans
33 c2odlA_
not modelled
7.8
25
PDB header: cell adhesionChain: A: PDB Molecule: adhesin;PDBTitle: crystal structure of the hmw1 secretion domain from2 haemophilus influenzae
34 c3on9B_
not modelled
7.8
33
PDB header: viral proteinChain: B: PDB Molecule: tumour necrosis factor receptor;PDBTitle: the secret domain from ectromelia virus
35 c2w91A_
not modelled
7.6
18
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-n-acetylglucosaminidase d;PDBTitle: structure of a streptococcus pneumoniae family 85 glycoside2 hydrolase, endo-d.
36 c2uygF_
not modelled
7.6
23
PDB header: lyaseChain: F: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
37 d1h05a_
not modelled
7.5
23
Fold: Flavodoxin-likeSuperfamily: Type II 3-dehydroquinate dehydrataseFamily: Type II 3-dehydroquinate dehydratase38 d1vi0a1
not modelled
7.5
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain39 c2rngA_
not modelled
7.5
100
PDB header: antimicrobial proteinChain: A: PDB Molecule: big defensin;PDBTitle: solution structure of big defensin
40 d1gmua2
not modelled
7.4
47
Fold: Ferredoxin-likeSuperfamily: Urease metallochaperone UreE, C-terminal domainFamily: Urease metallochaperone UreE, C-terminal domain41 d2p7ja1
not modelled
7.3
13
Fold: Profilin-likeSuperfamily: Sensory domain-likeFamily: YkuI C-terminal domain-like42 d1s57a_
not modelled
7.3
17
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK43 d2k49a1
not modelled
7.3
25
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like44 c3n8kG_
not modelled
6.9
23
PDB header: lyaseChain: G: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: type ii dehydroquinase from mycobacterium tuberculosis complexed with2 citrazinic acid
45 d1h3ga1
not modelled
6.8
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes46 d2fm8c1
not modelled
6.8
47
Fold: SipA N-terminal domain-likeSuperfamily: SipA N-terminal domain-likeFamily: SipA N-terminal domain-like47 d1mzha_
not modelled
6.8
18
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase48 c3gioA_
not modelled
6.6
45
PDB header: dna binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the tnf-alpha inducing protein (tip2 alpha) from helicobacter pylori
49 d2jmua1
not modelled
6.6
45
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain50 c3e0zB_
not modelled
6.6
23
PDB header: unknown functionChain: B: PDB Molecule: protein of unknown function;PDBTitle: crystal structure of a putative imidazole glycerol phosphate synthase2 homolog (eubrec_1070) from eubacterium rectale at 1.75 a resolution
51 c3gdbA_
not modelled
6.6
17
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein spr0440;PDBTitle: crystal structure of spr0440 glycoside hydrolase domain,2 endo-d from streptococcus pneumoniae r6
52 d1sg5a1
not modelled
6.5
20
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: Rof-like53 d1bdga2
not modelled
6.5
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase54 c1qhaA_
not modelled
6.4
24
PDB header: transferaseChain: A: PDB Molecule: protein (hexokinase);PDBTitle: human hexokinase type i complexed with atp analogue amp-pnp
55 c2po4A_
not modelled
6.4
29
PDB header: transferaseChain: A: PDB Molecule: virion rna polymerase;PDBTitle: x-ray crystal structure of polymerase domain of the2 bacteriophage n4 virion rna polymerase
56 c2l5rA_
not modelled
6.3
100
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide alyteserin-1c;PDBTitle: conformational and membrane interactins studies of antimicrobial2 peptide alyteserin-1c
57 c1s1iA_
not modelled
6.3
15
PDB header: ribosomeChain: A: PDB Molecule: 60s ribosomal protein l1;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
58 c2zkr5_
not modelled
6.3
33
PDB header: ribosomal protein/rnaChain: 5: PDB Molecule: 60s ribosomal protein l10a;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
59 d1eara2
not modelled
6.3
27
Fold: Ferredoxin-likeSuperfamily: Urease metallochaperone UreE, C-terminal domainFamily: Urease metallochaperone UreE, C-terminal domain60 d1lmea_
not modelled
6.2
26
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase61 d1g47a2
not modelled
6.2
32
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain62 d3bbba1
not modelled
6.2
15
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK63 c3sojA_
not modelled
6.1
58
PDB header: cell adhesionChain: A: PDB Molecule: pile;PDBTitle: francisella tularensis pilin pile
64 c2w3tA_
not modelled
6.0
19
PDB header: hydrolaseChain: A: PDB Molecule: peptide deformylase;PDBTitle: chloro complex of the ni-form of e.coli deformylase
65 d2az3a1
not modelled
6.0
17
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK66 c3dgpA_
not modelled
6.0
30
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase ii transcription factor b subunit 2;PDBTitle: crystal structure of the complex between tfb5 and the c-terminal2 domain of tfb2
67 d1gtza_
not modelled
5.8
40
Fold: Flavodoxin-likeSuperfamily: Type II 3-dehydroquinate dehydrataseFamily: Type II 3-dehydroquinate dehydratase68 d1c6ra_
not modelled
5.8
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c69 d1txla_
not modelled
5.8
21
Fold: LipocalinsSuperfamily: LipocalinsFamily: Hypothetical protein YodA70 c1txlA_
not modelled
5.8
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: metal-binding protein yoda;PDBTitle: crystal structure of metal-binding protein yoda from e.2 coli, pfam duf149
71 d3bl2a1
not modelled
5.7
31
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death72 d1u7na_
not modelled
5.7
18
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: PlsX-like73 d1pd0a1
not modelled
5.7
27
Fold: ERP29 C domain-likeSuperfamily: Helical domain of Sec23/24Family: Helical domain of Sec23/2474 d1rlra1
not modelled
5.7
16
Fold: R1 subunit of ribonucleotide reductase, N-terminal domainSuperfamily: R1 subunit of ribonucleotide reductase, N-terminal domainFamily: R1 subunit of ribonucleotide reductase, N-terminal domain75 c1v4sA_
not modelled
5.6
22
PDB header: transferaseChain: A: PDB Molecule: glucokinase isoform 2;PDBTitle: crystal structure of human glucokinase
76 d3c07a1
not modelled
5.6
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain77 d2c4va1
not modelled
5.6
25
Fold: Flavodoxin-likeSuperfamily: Type II 3-dehydroquinate dehydrataseFamily: Type II 3-dehydroquinate dehydratase78 c2i5nH_
not modelled
5.6
40
PDB header: photosynthesisChain: H: PDB Molecule: reaction center protein h chain;PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
79 d1rzhh1
not modelled
5.5
53
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: Photosynthetic reaction centre, H-chain, cytoplasmic domain80 c1eysH_
not modelled
5.5
24
PDB header: electron transportChain: H: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
81 c3i3lA_
not modelled
5.5
37
PDB header: hydrolaseChain: A: PDB Molecule: alkylhalidase cmls;PDBTitle: crystal structure of cmls, a flavin-dependent halogenase
82 c2ew7A_
not modelled
5.5
23
PDB header: hydrolaseChain: A: PDB Molecule: peptide deformylase;PDBTitle: crystal structure of helicobacter pylori peptide deformylase
83 d1ufya_
not modelled
5.5
16
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase84 d1wlfa1
not modelled
5.5
63
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like85 c3h43F_
not modelled
5.5
33
PDB header: hydrolaseChain: F: PDB Molecule: proteasome-activating nucleotidase;PDBTitle: n-terminal domain of the proteasome-activating nucleotidase2 of methanocaldococcus jannaschii
86 c3c8iA_
not modelled
5.5
27
PDB header: membrane proteinChain: A: PDB Molecule: putative membrane protein;PDBTitle: crystal structure of a putative membrane protein from corynebacterium2 diphtheriae
87 c3fk2B_
not modelled
5.4
13
PDB header: signaling protein, hydrolase activatorChain: B: PDB Molecule: glucocorticoid receptor dna-binding factor 1;PDBTitle: crystal structure of the rhogap domain of human2 glucocorticoid receptor dna-binding factor 1
88 d2ctda1
not modelled
5.4
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H289 c3kboB_
not modelled
5.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxylate/hydroxypyruvate reductase a;PDBTitle: 2.14 angstrom crystal structure of putative oxidoreductase (ycdw) from2 salmonella typhimurium in complex with nadp
90 d1t33a1
not modelled
5.4
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain91 d1v0ea2
not modelled
5.3
40
Fold: Triple-stranded beta-helixSuperfamily: Phage fibre proteinsFamily: Endo-alpha-sialidase92 d1l5ja3
not modelled
5.3
17
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain93 c3fn2A_
not modelled
5.3
22
PDB header: transferaseChain: A: PDB Molecule: putative sensor histidine kinase domain;PDBTitle: crystal structure of a putative sensor histidine kinase domain from2 clostridium symbiosum atcc 14940
94 d1gqoa_
not modelled
5.3
27
Fold: Flavodoxin-likeSuperfamily: Type II 3-dehydroquinate dehydrataseFamily: Type II 3-dehydroquinate dehydratase95 d1dwua_
not modelled
5.3
23
Fold: Ribosomal protein L1Superfamily: Ribosomal protein L1Family: Ribosomal protein L196 d2fbqa1
not modelled
5.3
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain97 d1ix1a_
not modelled
5.3
23
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase98 d1fqva1
not modelled
5.2
55
Fold: F-box domainSuperfamily: F-box domainFamily: F-box domain99 c3gnzP_
not modelled
5.2
22
PDB header: toxinChain: P: PDB Molecule: 25 kda protein elicitor;PDBTitle: toxin fold for microbial attack and plant defense