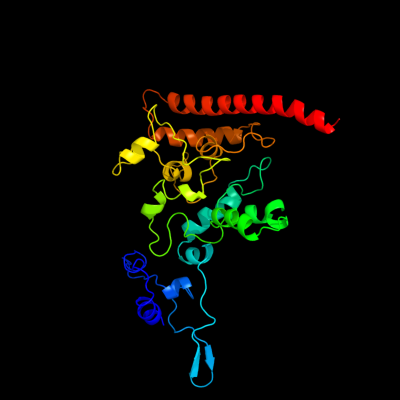
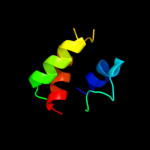
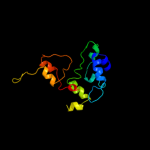
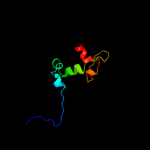
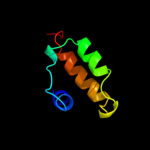
1 d1fgja_
99.9
17
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif2 c1fgjA_
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxylamine oxidoreductase;PDBTitle: x-ray structure of hydroxylamine oxidoreductase
3 c2j7aC_
99.9
27
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c quinol dehydrogenase nrfh;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
4 d2rdza1
98.6
21
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif5 c2bpbB_
98.6
17
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
6 d1sp3a_
98.5
18
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif7 d1fs7a_
98.4
17
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif8 c1fs9A_
98.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase;PDBTitle: cytochrome c nitrite reductase from wolinella succinogenes-azide2 complex
9 c1oahA_
98.4
16
PDB header: reductaseChain: A: PDB Molecule: cytochrome c nitrite reductase;PDBTitle: cytochrome c nitrite reductase from desulfovibrio2 desulfuricans atcc 27774: the relevance of the two3 calcium sites in the structure of the catalytic subunit4 (nrfa).
10 d1oaha_
98.4
16
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif11 c2p0bA_
98.2
19
PDB header: electron transportChain: A: PDB Molecule: cytochrome c-type protein nrfb;PDBTitle: crystal structure of chemically-reduced e.coli nrfb
12 d1pbya1
98.0
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 213 d1jmxa1
97.9
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 214 c3f29A_
97.8
22
PDB header: oxidoreductaseChain: A: PDB Molecule: eight-heme nitrite reductase;PDBTitle: structure of the thioalkalivibrio nitratireducens2 cytochrome c nitrite reductase in complex with sulfite
15 c2vr0A_
97.7
28
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase, catalytic subunit nfra;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex bound to the hqno inhibitor
16 c2fwtA_
97.7
24
PDB header: electron transportChain: A: PDB Molecule: dhc, diheme cytochrome c;PDBTitle: crystal structure of dhc purified from rhodobacter2 sphaeroides
17 c2j7aE_
97.6
29
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c nitrite reductase nrfa;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
18 d1qdba_
97.6
24
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif19 c3pmqA_
97.5
20
PDB header: electron transportChain: A: PDB Molecule: decaheme cytochrome c mtrf;PDBTitle: crystal structure of the outer membrane decaheme cytochrome mtrf
20 d1ft5a_
97.5
18
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif21 c3oueA_
not modelled
97.0
18
PDB header: electron transportChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: structure of c-terminal hexaheme fragment of gsu1996
22 d1mz4a_
not modelled
96.8
15
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c23 c2a3mA_
not modelled
96.7
35
PDB header: electron transportChain: A: PDB Molecule: cog3005: nitrate/tmao reductases, membrane-bound tetrahemePDBTitle: structure of desulfovibrio desulfuricans g20 tetraheme cytochrome2 (oxidized form)
24 c1pbyA_
not modelled
96.6
22
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein amine dehydrogenase 60 kdaPDBTitle: structure of the phenylhydrazine adduct of the2 quinohemoprotein amine dehydrogenase from paracoccus3 denitrificans at 1.7 a resolution
25 c1w5cT_
not modelled
96.5
15
PDB header: photosynthesisChain: T: PDB Molecule: cytochrome c-550;PDBTitle: photosystem ii from thermosynechococcus elongatus
26 c3ouqA_
not modelled
96.5
21
PDB header: electron transportChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: structure of n-terminal hexaheme fragment of gsu1996
27 d1f1ca_
not modelled
96.4
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c28 c1kb0A_
not modelled
96.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein alcohol dehydrogenase;PDBTitle: crystal structure of quinohemoprotein alcohol dehydrogenase from2 comamonas testosteroni
29 d1h9xa1
not modelled
96.2
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase30 c1jrxA_
not modelled
96.2
29
PDB header: oxidoreductaseChain: A: PDB Molecule: flavocytochrome c;PDBTitle: crystal structure of arg402ala mutant flavocytochrome c32 from shewanella frigidimarina
31 c3cp5A_
not modelled
96.2
25
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: cytochrome c from rhodothermus marinus
32 d1e29a_
not modelled
96.2
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c33 c2zonG_
not modelled
96.2
34
PDB header: oxidoreductase/electron transportChain: G: PDB Molecule: cytochrome c551;PDBTitle: crystal structure of electron transfer complex of nitrite2 reductase with cytochrome c
34 d1nira1
not modelled
96.1
19
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase35 c2e84A_
not modelled
96.0
18
PDB header: electron transportChain: A: PDB Molecule: high-molecular-weight cytochrome c;PDBTitle: crystal structure of high-molecular weight cytochrome c2 from desulfovibrio vulgaris (miyazaki f) in the presence3 of zinc ion
36 d1e2rb1
not modelled
96.0
23
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase37 d1i77a_
not modelled
95.9
36
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like38 d1d4ca1
not modelled
95.9
24
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif39 c1jmxA_
not modelled
95.8
27
PDB header: oxidoreductaseChain: A: PDB Molecule: amine dehydrogenase;PDBTitle: crystal structure of a quinohemoprotein amine dehydrogenase2 from pseudomonas putida
40 d1wada_
not modelled
95.8
26
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like41 c1qo8A_
not modelled
95.8
19
PDB header: oxidoreductaseChain: A: PDB Molecule: flavocytochrome c3 fumarate reductase;PDBTitle: the structure of the open conformation of a flavocytochrome2 c3 fumarate reductase
42 d1qo8a1
not modelled
95.7
20
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif43 c1kv9A_
not modelled
95.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: type ii quinohemoprotein alcohol dehydrogenase;PDBTitle: structure at 1.9 a resolution of a quinohemoprotein alcohol2 dehydrogenase from pseudomonas putida hk5
44 c2xtsD_
not modelled
95.7
25
PDB header: oxidoreductase/electron transportChain: D: PDB Molecule: cytochrome;PDBTitle: crystal structure of the sulfane dehydrogenase soxcd from paracoccus2 pantotrophus
45 d1j0pa_
not modelled
95.6
34
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like46 c3a9fA_
not modelled
95.5
22
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crystal structure of the c-terminal domain of cytochrome cz2 from chlorobium tepidum
47 d1hzua1
not modelled
95.4
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase48 d1kv9a1
not modelled
95.4
19
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinoprotein alcohol dehydrogenase, C-terminal domain49 d1ls9a_
not modelled
95.4
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c50 d2i5nc1
not modelled
95.4
14
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Photosynthetic reaction centre (cytochrome subunit)51 c2jblC_
not modelled
95.4
14
PDB header: electron transportChain: C: PDB Molecule: photosynthetic reaction center cytochrome cPDBTitle: photosynthetic reaction center from blastochloris viridis
52 c3dmiA_
not modelled
95.3
30
PDB header: electron transportChain: A: PDB Molecule: cytochrome c6;PDBTitle: crystallization and structural analysis of cytochrome c62 from the diatom phaeodactylum tricornutum at 1.5 a3 resolution
53 d2ctha_
not modelled
95.2
35
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like54 d1wvec1
not modelled
95.2
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c55 d1kb0a1
not modelled
95.1
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinoprotein alcohol dehydrogenase, C-terminal domain56 d1qksa1
not modelled
95.1
26
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase57 d1ytca_
not modelled
95.0
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c58 c2zooA_
not modelled
94.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nitrite reductase;PDBTitle: crystal structure of nitrite reductase from pseudoalteromonas2 haloplanktis tac125
59 d1ccra_
not modelled
94.9
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c60 c2l4dA_
not modelled
94.9
18
PDB header: electron transportChain: A: PDB Molecule: sco1/senc family protein/cytochrome c;PDBTitle: cytochrome c domain of pp3183 protein from pseudomonas putida
61 d1yeba_
not modelled
94.8
21
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c62 d1fcdc1
not modelled
94.8
27
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c63 d1ycca_
not modelled
94.8
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c64 c1nnoA_
not modelled
94.8
22
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrite reductase;PDBTitle: conformational changes occurring upon no binding in nitrite2 reductase from pseudomonas aeruginosa
65 d1up9a_
not modelled
94.7
31
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like66 d2gc4d1
not modelled
94.7
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c67 d1ofwa_
not modelled
94.7
27
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like68 d1c75a_
not modelled
94.7
17
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c69 d1cyja_
not modelled
94.7
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c70 d1hj3a1
not modelled
94.7
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase71 d1lfma_
not modelled
94.7
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c72 c2d0wA_
not modelled
94.6
18
PDB header: electron transportChain: A: PDB Molecule: cytochrome cl;PDBTitle: crystal structure of cytochrome cl from hyphomicrobium2 denitrificans
73 d1dy7b1
not modelled
94.6
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase74 d1wejf_
not modelled
94.6
19
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c75 c3dr0B_
not modelled
94.5
22
PDB header: electron transportChain: B: PDB Molecule: cytochrome c6;PDBTitle: structure of reduced cytochrome c6 from synechococcus sp.2 pcc 7002
76 d2cvca1
not modelled
94.5
22
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like77 d1duwa_
not modelled
94.5
22
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like78 c2w9kA_
not modelled
94.5
27
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crithidia fasciculata cytochrome c
79 d1ctja_
not modelled
94.4
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c80 c2cvcA_
not modelled
94.4
23
PDB header: electron transportChain: A: PDB Molecule: high-molecular-weight cytochrome c precursor;PDBTitle: crystal structure of high-molecular weight cytochrome c2 from desulfovibrio vulgaris (hildenborough)
81 d1c52a_
not modelled
94.4
23
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c82 d2cy3a_
not modelled
94.4
21
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like83 d1f1fa_
not modelled
94.4
21
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c84 d1gyoa_
not modelled
94.1
32
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like85 d1hroa_
not modelled
94.1
16
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c86 d1y0pa1
not modelled
94.1
22
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif87 c1w2lA_
not modelled
94.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome oxidase subunit ii;PDBTitle: cytochrome c domain of caa3 oxygen oxidoreductase
88 d1cc5a_
not modelled
94.1
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c89 d1fcdc2
not modelled
94.0
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c90 d1j3sa_
not modelled
94.0
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c91 c2v07A_
not modelled
94.0
22
PDB header: photosynthesisChain: A: PDB Molecule: cytochrome c6;PDBTitle: structure of the arabidopsis thaliana cytochrome c6a v52q2 variant
92 c2zxyA_
not modelled
94.0
17
PDB header: oxygen binding, transport proteinChain: A: PDB Molecule: cytochrome c552;PDBTitle: crystal structure of cytochrome c555 from aquifex aeolicus
93 d2c8sa1
not modelled
93.9
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c94 d1co6a_
not modelled
93.9
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c95 d1aqea_
not modelled
93.9
18
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like96 c3o0rC_
not modelled
93.8
24
PDB header: immune system/oxidoreductaseChain: C: PDB Molecule: nitric oxide reductase subunit c;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
97 c2zzsW_
not modelled
93.8
26
PDB header: electron transportChain: W: PDB Molecule: PDBTitle: crystal structure of cytochrome c554 from vibrio2 parahaemolyticus strain rimd2210633
98 d1cnoa_
not modelled
93.8
28
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c99 d1kx7a_
not modelled
93.7
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c100 c3cu4A_
not modelled
93.7
28
PDB header: electron transportChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: omcf, outer membrance cytochrome f from geobacter2 sulfurreducens
101 d1m1qa_
not modelled
93.6
34
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif102 d1gdva_
not modelled
93.6
21
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c103 d1h1oa2
not modelled
93.5
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c104 d1m70a1
not modelled
93.3
26
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c105 d1c6ra_
not modelled
93.3
17
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c106 d1c53a_
not modelled
93.1
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c107 c1eysC_
not modelled
92.9
22
PDB header: electron transportChain: C: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
108 d1eysc_
not modelled
92.9
22
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Photosynthetic reaction centre (cytochrome subunit)109 d19hca_
not modelled
92.5
27
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like110 c1z1nX_
not modelled
92.3
17
PDB header: electron transportChain: X: PDB Molecule: sixteen heme cytochrome;PDBTitle: crystal structure of the sixteen heme cytochrome from desulfovibrio2 gigas
111 d1c6sa_
not modelled
92.0
17
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c112 d1m70a2
not modelled
92.0
28
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c113 d1dvha_
not modelled
91.8
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c114 c1gq1B_
not modelled
91.8
22
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome cd1 nitrite reductase;PDBTitle: cytochrome cd1 nitrite reductase, y25s mutant, oxidised2 form
115 d1gksa_
not modelled
91.8
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c116 c1yiqA_
not modelled
91.2
22
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein alcohol dehydrogenase;PDBTitle: molecular cloning and structural analysis of2 quinohemoprotein alcohol dehydrogenase adhiig from3 pseudomonas putida hk5. compariison to the other4 quinohemoprotein alcohol dehydrogenase adhiib found in the5 same microorganism.
117 c1fcdD_
not modelled
91.0
21
PDB header: electron transport(flavocytochrome)Chain: D: PDB Molecule: flavocytochrome c sulfide dehydrogenasePDBTitle: the structure of flavocytochrome c sulfide dehydrogenase2 from a purple phototrophic bacterium chromatium vinosum at3 2.5 angstroms resolution
118 d1qn2a_
not modelled
90.9
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c119 c2d0sA_
not modelled
90.6
31
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crystal structure of the cytochrome c552 from moderate2 thermophilic bacterium, hydrogenophilus thermoluteolus
120 c3m97X_
not modelled
90.4
17
PDB header: electron transportChain: X: PDB Molecule: cytochrome c-552;PDBTitle: structure of the soluble domain of cytochrome c552 with its flexible2 linker segment from paracoccus denitrificans