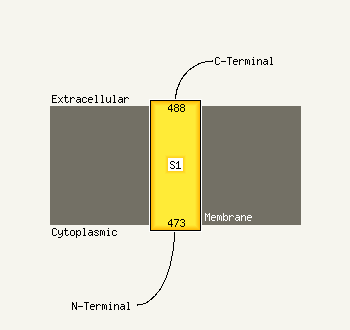
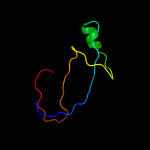
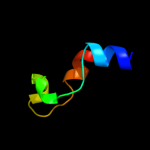
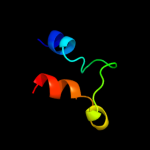
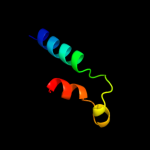
1 c2qzaA_
95.3
27
PDB header: ligaseChain: A: PDB Molecule: secreted effector protein;PDBTitle: crystal structure of salmonella effector protein sopa
2 c3nb2B_
94.6
31
PDB header: ligaseChain: B: PDB Molecule: secreted effector protein;PDBTitle: crystal structure of e. coli o157:h7 effector protein nlel
3 d1qcsa2
24.9
21
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like4 d1vmja_
19.3
24
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like5 c1ve0A_
18.3
29
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein (st2072);PDBTitle: crystal structure of uncharacterized protein st2072 from sulfolobus2 tokodaii
6 d1vpha_
18.0
24
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like7 c2w8dB_
17.4
17
PDB header: transferaseChain: B: PDB Molecule: processed glycerol phosphate lipoteichoic acid synthase 2;PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
8 c3ik5A_
16.9
33
PDB header: viral protein/signaling proteinChain: A: PDB Molecule: protein nef;PDBTitle: sivmac239 nef in complex with tcr zeta itam 1 polypeptide2 (a63-r80)
9 d1s4ka_
16.6
24
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: YdiL-like10 c2p6cB_
15.9
28
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: aq_2013 protein;PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
11 d2py5a1
14.9
33
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: DnaQ-like 3'-5' exonuclease12 c2jynA_
14.2
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0368 protein ypl225w;PDBTitle: a novel solution nmr structure of protein yst0336 from2 saccharomyces cerevisiae. northeast structural genomics3 consortium target yt51/ontario centre for structural4 proteomics target yst0336
13 c2xi1A_
14.2
39
PDB header: viral proteinChain: A: PDB Molecule: nef;PDBTitle: crystal structure of the hiv-1 nef sequenced from a patient's sample
14 d1efnb_
13.4
33
Fold: Regulatory factor NefSuperfamily: Regulatory factor NefFamily: Regulatory factor Nef15 c3rbbA_
12.4
39
PDB header: viral protein, protein bindingChain: A: PDB Molecule: protein nef;PDBTitle: hiv-1 nef protein in complex with engineered hck sh3 domain
16 d2nefa_
12.1
33
Fold: Regulatory factor NefSuperfamily: Regulatory factor NefFamily: Regulatory factor Nef17 c3ebcA_
11.9
62
PDB header: hydrolase/dnaChain: A: PDB Molecule: type-2 restriction enzyme hincii;PDBTitle: structure of n141a hincii with cognate dna
18 d1bf4a_
11.8
42
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea19 c1p82A_
11.6
62
PDB header: chaperoneChain: A: PDB Molecule: 10 kda chaperonin;PDBTitle: nmr structure of 1-25 fragment of mycobacterium2 tuberculosis cpn10
20 d2pgca1
11.5
56
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Marine metagenome family DABB321 d1tx3a1
not modelled
11.3
62
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease HincII22 d1vj0a1
not modelled
10.9
20
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain23 c2xrhA_
not modelled
10.8
32
PDB header: unknown functionChain: A: PDB Molecule: protein hp0721;PDBTitle: crystal structure of the truncated form of hp0721
24 c2elpA_
not modelled
9.0
55
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
25 d1alqa_
not modelled
8.9
22
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase26 d1ug7a_
not modelled
8.6
20
Fold: Four-helical up-and-down bundleSuperfamily: Domain from hypothetical 2610208m17rik proteinFamily: Domain from hypothetical 2610208m17rik protein27 c2qyxB_
not modelled
8.3
31
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein mj0159;PDBTitle: crystal structure of uncharacterized protein mj0159 from2 methanocaldococcus jannaschii
28 c2jvfA_
not modelled
8.1
32
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
29 c1qysA_
not modelled
7.9
29
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
30 c2ejsA_
not modelled
7.9
17
PDB header: ligaseChain: A: PDB Molecule: autocrine motility factor receptor, isoform 2;PDBTitle: solution structure of ruh-076, a human cue domain
31 d1y14a_
not modelled
7.8
12
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: RNA polymerase II subunit RBP4 (RpoF)32 c2r8rB_
not modelled
7.7
26
PDB header: transferaseChain: B: PDB Molecule: sensor protein;PDBTitle: crystal structure of the n-terminal region (19..243) of sensor protein2 kdpd from pseudomonas syringae pv. tomato str. dc3000
33 c3lgeG_
not modelled
7.3
47
PDB header: lyase/protein bindingChain: G: PDB Molecule: sorting nexin-9;PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
34 c3lgeF_
not modelled
7.3
47
PDB header: lyase/protein bindingChain: F: PDB Molecule: sorting nexin-9;PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
35 c3h0gP_
not modelled
6.9
21
PDB header: transcriptionChain: P: PDB Molecule: dna-directed rna polymerase ii subunit rpb4;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
36 c3l76B_
not modelled
6.7
42
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
37 c1qdnA_
not modelled
6.6
21
PDB header: fusion proteinChain: A: PDB Molecule: protein (n-ethylmaleimide sensitive fusionPDBTitle: amino terminal domain of the n-ethylmaleimide sensitive2 fusion protein (nsf)
38 c2klqA_
not modelled
6.5
29
PDB header: replicationChain: A: PDB Molecule: dna replication licensing factor mcm6;PDBTitle: the solution structure of cbd of human mcm6
39 d1vmfa_
not modelled
6.1
23
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like40 d2qrdb1
not modelled
6.0
44
Fold: AMPKBI-likeSuperfamily: AMPKBI-likeFamily: AMPKBI-like41 d1p2fa1
not modelled
5.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like42 c3fo8D_
not modelled
5.9
43
PDB header: viral proteinChain: D: PDB Molecule: tail sheath protein gp18;PDBTitle: crystal structure of the bacteriophage t4 tail sheath2 protein, protease resistant fragment gp18pr
43 c2ex3I_
not modelled
5.9
33
PDB header: transferase/replicationChain: I: PDB Molecule: dna polymerase;PDBTitle: bacteriophage phi29 dna polymerase bound to terminal protein
44 c2wzpR_
not modelled
5.8
28
PDB header: viral proteinChain: R: PDB Molecule: lactococcal phage p2 orf16;PDBTitle: structures of lactococcal phage p2 baseplate shed light on2 a novel mechanism of host attachment and activation in3 siphoviridae
45 c3gpkA_
not modelled
5.8
12
PDB header: isomeraseChain: A: PDB Molecule: ppic-type peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
46 c2k0mA_
not modelled
5.7
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of the uncharacterized protein from2 rhodospirillum rubrum gene locus rru_a0810. northeast3 structural genomics target rrr43
47 d1gado1
not modelled
5.6
58
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain48 d1ggaa1
not modelled
5.6
58
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain49 d2qlvb2
not modelled
5.5
44
Fold: AMPKBI-likeSuperfamily: AMPKBI-likeFamily: AMPKBI-like50 d2nn4a1
not modelled
5.4
37
Fold: YqgQ-likeSuperfamily: YqgQ-likeFamily: YqgQ-like51 c3jszA_
not modelled
5.4
35
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: legionella pneumophila glucosyltransferase lgt1 n293a with udp-glc
52 d2v8qb1
not modelled
5.3
44
Fold: AMPKBI-likeSuperfamily: AMPKBI-likeFamily: AMPKBI-like53 c2qdjA_
not modelled
5.2
11
PDB header: antitumor proteinChain: A: PDB Molecule: retinoblastoma-associated protein;PDBTitle: crystal structure of the retinoblastoma protein n-domain2 provides insight into tumor suppression, ligand3 interaction and holoprotein architecture
54 c2cp8A_
not modelled
5.2
31
PDB header: protein bindingChain: A: PDB Molecule: next to brca1 gene 1 protein;PDBTitle: solution structure of the rsgi ruh-046, a uba domain from2 human next to brca1 gene 1 protein (kiaa0049 protein)3 r923h variant
55 c2cu5C_
not modelled
5.2
27
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: conserved hypothetical protein tt1486;PDBTitle: crystal structure of the conserved hypothetical protein tt1486 from2 thermus thermophilus hb8
56 c1zk6A_
not modelled
5.1
38
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: nmr solution structure of b. subtilis prsa ppiase