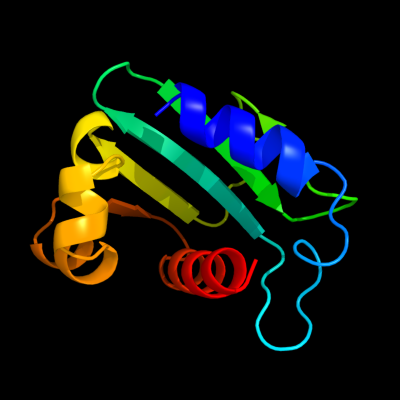
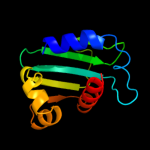
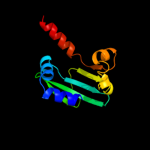
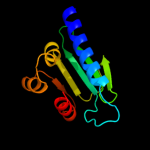
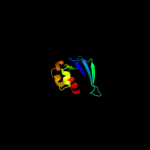
1 d1ikta_
98.8
15
Fold: SCP-likeSuperfamily: SCP-likeFamily: Sterol carrier protein, SCP2 c3bn8A_
98.8
12
PDB header: transport proteinChain: A: PDB Molecule: putative sterol carrier protein 2;PDBTitle: crystal structure of a putative sterol carrier protein type 2 (af1534)2 from archaeoglobus fulgidus dsm 4304 at 2.11 a resolution
3 c3bdqB_
98.8
16
PDB header: lipid transportChain: B: PDB Molecule: sterol carrier protein 2-like 2;PDBTitle: room tempreture crystal structure of sterol carrier protein-2 2 like-2
4 d1c44a_
98.6
16
Fold: SCP-likeSuperfamily: SCP-likeFamily: Sterol carrier protein, SCP5 d1pz4a_
98.4
10
Fold: SCP-likeSuperfamily: SCP-likeFamily: Sterol carrier protein, SCP6 d2cfua1
98.4
15
Fold: SCP-likeSuperfamily: SCP-likeFamily: Alkylsulfatase C-terminal domain-like7 c3bkrA_
98.2
18
PDB header: lipid binding proteinChain: A: PDB Molecule: sterol carrier protein-2 like-3;PDBTitle: crystal structure of sterol carrier protein-2 like-3 (scp2-2 l3) from aedes aegypti
8 d1wfra_
97.9
19
Fold: SCP-likeSuperfamily: SCP-likeFamily: Sterol carrier protein, SCP9 c2cfuA_
97.1
15
PDB header: hydrolaseChain: A: PDB Molecule: sdsa1;PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
10 d2i00a1
92.7
13
Fold: SCP-likeSuperfamily: SCP-likeFamily: EF1021 C-terminal domain-like11 d2hv2a1
91.2
16
Fold: SCP-likeSuperfamily: SCP-likeFamily: EF1021 C-terminal domain-like12 c2i00D_
86.3
13
PDB header: transferaseChain: D: PDB Molecule: acetyltransferase, gnat family;PDBTitle: crystal structure of acetyltransferase (gnat family) from enterococcus2 faecalis
13 c2wukD_
66.9
22
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
14 d2ozga1
61.2
18
Fold: SCP-likeSuperfamily: SCP-likeFamily: EF1021 C-terminal domain-like15 c1aq5C_
58.4
35
PDB header: coiled-coilChain: C: PDB Molecule: cartilage matrix protein;PDBTitle: high-resolution solution nmr structure of the trimeric coiled-coil2 domain of chicken cartilage matrix protein, 20 structures
16 c1m1jA_
56.1
15
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha subunit;PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
17 c2hv2D_
56.0
17
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of conserved protein of unknown function from2 enterococcus faecalis v583 at 2.4 a resolution, probable n-3 acyltransferase
18 c2xdjF_
54.5
3
PDB header: unknown functionChain: F: PDB Molecule: uncharacterized protein ybgf;PDBTitle: crystal structure of the n-terminal domain of e.coli ybgf
19 c1t3jA_
51.2
22
PDB header: membrane proteinChain: A: PDB Molecule: mitofusin 1;PDBTitle: mitofusin domain hr2 v686m/i708m mutant
20 c1go4F_
46.3
15
PDB header: cell cycleChain: F: PDB Molecule: mad1 (mitotic arrest deficient)-like 1;PDBTitle: crystal structure of mad1-mad2 reveals a conserved mad22 binding motif in mad1 and cdc20.
21 c1dipA_
not modelled
43.4
24
PDB header: acetylationChain: A: PDB Molecule: delta-sleep-inducing peptide immunoreactivePDBTitle: the solution structure of porcine delta-sleep-inducing2 peptide immunoreactive peptide, nmr, 10 structures
22 c2pnvA_
not modelled
42.7
20
PDB header: membrane proteinChain: A: PDB Molecule: small conductance calcium-activated potassiumPDBTitle: crystal structure of the leucine zipper domain of small-2 conductance ca2+-activated k+ (skca) channel from rattus3 norvegicus
23 c2yy0D_
not modelled
38.9
27
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
24 c1m7lA_
not modelled
35.7
29
PDB header: sugar binding proteinChain: A: PDB Molecule: pulmonary surfactant-associated protein d;PDBTitle: solution structure of the coiled-coil trimerization domain2 from lung surfactant protein d
25 c3n7zD_
not modelled
34.3
8
PDB header: transferaseChain: D: PDB Molecule: acetyltransferase, gnat family;PDBTitle: crystal structure of acetyltransferase from bacillus anthracis
26 c2akfC_
not modelled
34.0
29
PDB header: protein bindingChain: C: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
27 c2akfA_
not modelled
34.0
29
PDB header: protein bindingChain: A: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
28 c2akfB_
not modelled
34.0
29
PDB header: protein bindingChain: B: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
29 c3iqtA_
not modelled
29.8
10
PDB header: transferaseChain: A: PDB Molecule: signal transduction histidine-protein kinase bara;PDBTitle: structure of the hpt domain of sensor protein bara from escherichia2 coli cft073.
30 c1coiA_
not modelled
25.3
43
PDB header: alpha-helical bundleChain: A: PDB Molecule: coil-vald;PDBTitle: designed trimeric coiled coil-vald
31 c3rylB_
not modelled
25.2
13
PDB header: protein bindingChain: B: PDB Molecule: protein vpa1370;PDBTitle: dimerization domain of vibrio parahemolyticus vopl
32 c2ozgA_
not modelled
24.0
18
PDB header: transferaseChain: A: PDB Molecule: gcn5-related n-acetyltransferase;PDBTitle: crystal structure of gcn5-related n-acetyltransferase (yp_325469.1)2 from anabaena variabilis atcc 29413 at 2.00 a resolution
33 c3qh9A_
not modelled
24.0
20
PDB header: structural proteinChain: A: PDB Molecule: liprin-beta-2;PDBTitle: human liprin-beta2 coiled-coil
34 d1rf8b_
not modelled
23.1
13
Fold: Eukaryotic initiation factor 4f subunit eIF4g, eIF4e-binding domainSuperfamily: Eukaryotic initiation factor 4f subunit eIF4g, eIF4e-binding domainFamily: Eukaryotic initiation factor 4f subunit eIF4g, eIF4e-binding domain35 c3ghgD_
not modelled
21.7
9
PDB header: blood clottingChain: D: PDB Molecule: fibrinogen alpha chain;PDBTitle: crystal structure of human fibrinogen
36 c1lq0A_
not modelled
19.0
10
PDB header: hydrolaseChain: A: PDB Molecule: chitotriosidase;PDBTitle: crystal structure of human chitotriosidase at 2.2 angstrom2 resolution
37 c3gw6F_
not modelled
18.2
31
PDB header: chaperoneChain: F: PDB Molecule: endo-n-acetylneuraminidase;PDBTitle: intramolecular chaperone
38 d1wb0a1
not modelled
15.0
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase39 c3r1kA_
not modelled
15.0
17
PDB header: transferaseChain: A: PDB Molecule: enhanced intracellular survival protein;PDBTitle: crystal structure of acetyltransferase eis from mycobacterium2 tuberculosis h37rv in complex with coa and an acetamide moiety
40 c1ci6B_
not modelled
14.9
13
PDB header: transcriptionChain: B: PDB Molecule: transcription factor c/ebp beta;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
41 c3ojaB_
not modelled
14.6
10
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
42 c3p8cD_
not modelled
14.5
19
PDB header: protein bindingChain: D: PDB Molecule: wiskott-aldrich syndrome protein family member 1;PDBTitle: structure and control of the actin regulatory wave complex
43 c1hf9B_
not modelled
13.8
17
PDB header: atpase inhibitorChain: B: PDB Molecule: atpase inhibitor (mitochondrial);PDBTitle: c-terminal coiled-coil domain from bovine if1
44 c1gk7A_
not modelled
13.6
24
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 1a fragment (1a)
45 c1kddC_
not modelled
13.3
25
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
46 c1kddF_
not modelled
13.0
25
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
47 c1kddA_
not modelled
13.0
25
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
48 c2o1mB_
not modelled
12.8
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable amino-acid abc transporterPDBTitle: crystal structure of the probable amino-acid abc2 transporter extracellular-binding protein ytmk from3 bacillus subtilis. northeast structural genomics4 consortium target sr572
49 c3ctwB_
not modelled
12.7
14
PDB header: protein bindingChain: B: PDB Molecule: rcda;PDBTitle: crystal structure of rcda from caulobacter crescentus cb15
50 d1bgfa_
not modelled
12.6
7
Fold: Transcription factor STAT-4 N-domainSuperfamily: Transcription factor STAT-4 N-domainFamily: Transcription factor STAT-4 N-domain51 c1u0iA_
not modelled
12.6
43
PDB header: de novo proteinChain: A: PDB Molecule: iaal-e3;PDBTitle: iaal-e3/k3 heterodimer
52 c1wb0A_
not modelled
12.5
10
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: chitotriosidase 1;PDBTitle: specificity and affinity of natural product cyclopentapeptide2 inhibitor argifin against human chitinase
53 c3he4A_
not modelled
11.6
30
PDB header: de novo proteinChain: A: PDB Molecule: synzip6;PDBTitle: heterospecific coiled-coil pair synzip5:synzip6
54 d1rp3a2
not modelled
11.2
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain55 d1sxje1
not modelled
11.1
14
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain56 c3fxyA_
not modelled
10.4
8
PDB header: hydrolaseChain: A: PDB Molecule: acidic mammalian chitinase;PDBTitle: acidic mammalian chinase, catalytic domain
57 c2dt7A_
not modelled
10.1
26
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor 3a subunit 3;PDBTitle: solution structure of the second surp domain of human2 splicing factor sf3a120 in complex with a fragment of3 human splicing factor sf3a60
58 d1vf8a1
not modelled
9.6
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase59 d1kfta_
not modelled
9.5
19
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Excinuclease UvrC C-terminal domain60 c1kftA_
not modelled
9.5
19
PDB header: dna binding proteinChain: A: PDB Molecule: excinuclease abc subunit c;PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
61 c3iynR_
not modelled
9.2
14
PDB header: virusChain: R: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
62 c1kd9C_
not modelled
8.2
25
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
63 c1kd9A_
not modelled
8.2
25
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
64 c1kd9F_
not modelled
8.2
25
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
65 d1ivsa1
not modelled
7.9
19
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain66 d1st6a3
not modelled
7.8
16
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin67 c3myfB_
not modelled
7.7
9
PDB header: transferaseChain: B: PDB Molecule: sensor protein;PDBTitle: the crystal structure of the hpt domain from the hpt sensor hybrid2 histidine kinase from shewanella to 1.80a
68 c2l5gB_
not modelled
7.1
19
PDB header: transcription regulatorChain: B: PDB Molecule: putative uncharacterized protein ncor2;PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
69 c3t97B_
not modelled
6.8
6
PDB header: protein transportChain: B: PDB Molecule: nuclear pore complex protein nup54;PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup62/nup54
70 c2lgxA_
not modelled
6.5
20
PDB header: cell adhesionChain: A: PDB Molecule: fermitin family homolog 2;PDBTitle: nmr structure for kindle-2 n-terminus
71 c1jekA_
not modelled
6.5
15
PDB header: viral proteinChain: A: PDB Molecule: env polyprotein;PDBTitle: visna tm core structure
72 c1kd8F_
not modelled
6.4
29
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12ia16v;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
73 c1kd8A_
not modelled
6.4
29
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12ia16v;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
74 c1kd8C_
not modelled
6.4
29
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12ia16v;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
75 c3a7pB_
not modelled
6.3
8
PDB header: protein transportChain: B: PDB Molecule: autophagy protein 16;PDBTitle: the crystal structure of saccharomyces cerevisiae atg16
76 c3hugA_
not modelled
5.9
6
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
77 c3ljmB_
not modelled
5.5
41
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
78 c2jgoC_
not modelled
5.5
41
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l9c;PDBTitle: stucture of the arsenated de novo designed peptide coil ser2 l9c
79 c1cosB_
not modelled
5.4
41
PDB header: alpha-helical bundleChain: B: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
80 c1e9lA_
not modelled
5.3
10
PDB header: macrophage secretory proteinChain: A: PDB Molecule: ym1 secretory protein;PDBTitle: the crystal structure of novel mammalian lectin ym12 suggests a saccharide binding site
81 c2jgoB_
not modelled
5.3
39
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l9c;PDBTitle: stucture of the arsenated de novo designed peptide coil ser2 l9c
82 c3ljmC_
not modelled
5.3
39
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
83 c3ljmA_
not modelled
5.3
39
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
84 c2jgoA_
not modelled
5.3
39
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l9c;PDBTitle: stucture of the arsenated de novo designed peptide coil ser2 l9c
85 c1u2uA_
not modelled
5.3
32
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: nmr solution structure of a designed heterodimeric leucine2 zipper
86 d1ii5a_
not modelled
5.2
9
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like87 c1cosA_
not modelled
5.2
39
PDB header: alpha-helical bundleChain: A: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
88 c1cosC_
not modelled
5.2
39
PDB header: alpha-helical bundleChain: C: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
89 d1u00a1
not modelled
5.2
11
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Heat shock protein 70kD (HSP70), C-terminal subdomainFamily: Heat shock protein 70kD (HSP70), C-terminal subdomain90 c2xzrA_
not modelled
5.1
8
PDB header: cell adhesionChain: A: PDB Molecule: immunoglobulin-binding protein eibd;PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
91 d2hvma_
not modelled
5.1
32
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase