| 1 |
|
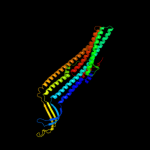
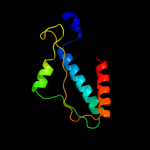
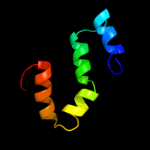
PDB 1tqq chain C
Region: 23 - 450
Aligned: 428
Modelled: 428
Confidence: 100.0%
Identity: 100%
PDB header:transport protein
Chain: C: PDB Molecule:outer membrane protein tolc;
PDBTitle: structure of tolc in complex with hexamminecobalt
Phyre2
| 2 |
|
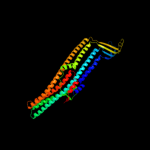
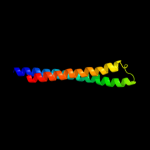
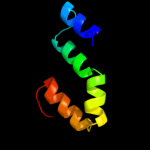
PDB 1ek9 chain A
Region: 23 - 449
Aligned: 427
Modelled: 427
Confidence: 100.0%
Identity: 99%
Fold: Outer membrane efflux proteins (OEP)
Superfamily: Outer membrane efflux proteins (OEP)
Family: Outer membrane efflux proteins (OEP)
Phyre2
| 3 |
|
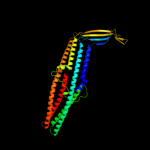
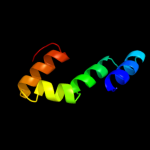
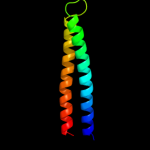
PDB 3pik chain A
Region: 24 - 427
Aligned: 393
Modelled: 404
Confidence: 100.0%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusc;
PDBTitle: outer membrane protein cusc
Phyre2
| 4 |
|
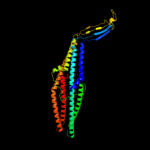
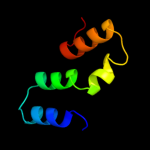
PDB 1wp1 chain A
Region: 24 - 433
Aligned: 400
Modelled: 410
Confidence: 100.0%
Identity: 21%
Fold: Outer membrane efflux proteins (OEP)
Superfamily: Outer membrane efflux proteins (OEP)
Family: Outer membrane efflux proteins (OEP)
Phyre2
| 5 |
|
PDB 1yc9 chain A
Region: 24 - 431
Aligned: 392
Modelled: 408
Confidence: 100.0%
Identity: 20%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein;
PDBTitle: the crystal structure of the outer membrane protein vcec from the2 bacterial pathogen vibrio cholerae at 1.8 resolution
Phyre2
| 6 |
|
PDB 2v7s chain A
Region: 235 - 331
Aligned: 97
Modelled: 97
Confidence: 51.5%
Identity: 10%
PDB header:unknown function
Chain: A: PDB Molecule:probable conserved lipoprotein lppa;
PDBTitle: crystal structure of the putative lipoprotein lppa from2 mycobacterium tuberculosis
Phyre2
| 7 |
|
PDB 3fpp chain B
Region: 343 - 425
Aligned: 83
Modelled: 83
Confidence: 37.6%
Identity: 8%
PDB header:membrane protein
Chain: B: PDB Molecule:macrolide-specific efflux protein maca;
PDBTitle: crystal structure of e.coli maca
Phyre2
| 8 |
|
PDB 2l06 chain A
Region: 157 - 212
Aligned: 56
Modelled: 56
Confidence: 26.3%
Identity: 11%
PDB header:protein binding
Chain: A: PDB Molecule:phycobilisome lcm core-membrane linker polypeptide;
PDBTitle: solution nmr structure of the pbs linker polypeptide domain (fragment2 254-400) of phycobilisome linker protein apce from synechocystis sp.3 pcc 6803. northeast structural genomics consortium target sgr209c
Phyre2
| 9 |
|
PDB 3ohw chain B
Region: 156 - 212
Aligned: 57
Modelled: 57
Confidence: 23.8%
Identity: 4%
PDB header:protein binding
Chain: B: PDB Molecule:phycobilisome lcm core-membrane linker polypeptide;
PDBTitle: x-ray structure of phycobilisome lcm core-membrane linker polypeptide2 (fragment 721-860) from synechocystis sp. pcc 6803, northeast3 structural genomics consortium target sgr209e
Phyre2
| 10 |
|
PDB 2ky4 chain A
Region: 156 - 212
Aligned: 57
Modelled: 57
Confidence: 21.2%
Identity: 9%
PDB header:photosynthesis
Chain: A: PDB Molecule:phycobilisome linker polypeptide;
PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome2 linker polypeptide from anabaena sp. northeast structural genomics3 consortium target nsr123e
Phyre2
| 11 |
|
PDB 2l3w chain A
Region: 159 - 212
Aligned: 54
Modelled: 54
Confidence: 20.1%
Identity: 11%
PDB header:photosynthesis
Chain: A: PDB Molecule:phycobilisome rod linker polypeptide;
PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome rod2 linker polypeptide from synechococcus elongatus, northeast structural3 genomics consortium target snr168a
Phyre2
| 12 |
|
PDB 3pru chain D
Region: 162 - 212
Aligned: 51
Modelled: 51
Confidence: 18.9%
Identity: 6%
PDB header:photosynthesis
Chain: D: PDB Molecule:phycobilisome 32.1 kda linker polypeptide, phycocyanin-
PDBTitle: crystal structure of phycobilisome 32.1 kda linker polypeptide,2 phycocyanin-associated, rod 1 (fragment 14-158) from synechocystis3 sp. pcc 6803, northeast structural genomics consortium target sgr182a
Phyre2
| 13 |
|
PDB 3sog chain A
Region: 313 - 427
Aligned: 113
Modelled: 115
Confidence: 17.1%
Identity: 7%
PDB header:structural protein
Chain: A: PDB Molecule:amphiphysin;
PDBTitle: crystal structure of the bar domain of human amphiphysin, isoform 1
Phyre2
| 14 |
|
PDB 1urq chain C
Region: 142 - 207
Aligned: 66
Modelled: 66
Confidence: 11.7%
Identity: 14%
PDB header:transport protein
Chain: C: PDB Molecule:synaptosomal-associated protein 25;
PDBTitle: crystal structure of neuronal q-snares in complex with2 r-snare motif of tomosyn
Phyre2
| 15 |
|
PDB 2z0v chain A
Region: 343 - 426
Aligned: 81
Modelled: 84
Confidence: 11.3%
Identity: 10%
PDB header:endocytosis
Chain: A: PDB Molecule:sh3-containing grb2-like protein 3;
PDBTitle: crystal structure of bar domain of endophilin-iii
Phyre2
| 16 |
|
PDB 2f1m chain A
Region: 350 - 422
Aligned: 73
Modelled: 73
Confidence: 9.2%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:acriflavine resistance protein a;
PDBTitle: conformational flexibility in the multidrug efflux system protein acra
Phyre2
| 17 |
|
PDB 3b5n chain C
Region: 147 - 207
Aligned: 61
Modelled: 61
Confidence: 7.8%
Identity: 13%
PDB header:membrane protein
Chain: C: PDB Molecule:protein transport protein sec9;
PDBTitle: structure of the yeast plasma membrane snare complex
Phyre2
| 18 |
|
PDB 1nhl chain A
Region: 156 - 207
Aligned: 52
Modelled: 52
Confidence: 5.4%
Identity: 13%
PDB header:protein transport
Chain: A: PDB Molecule:synaptosomal-associated protein 23;
PDBTitle: snap-23n structure
Phyre2