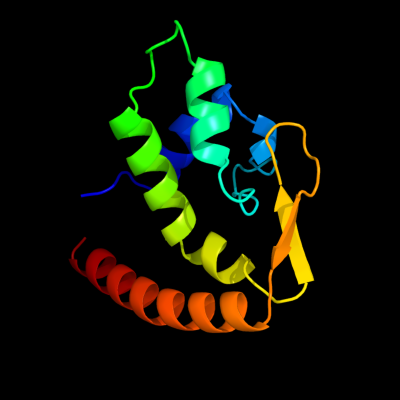
| 1 | d1dvoa_
|
|
 |
100.0 |
26 |
Fold:FinO-like
Superfamily:FinO-like
Family:FinO-like |
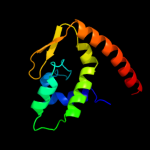
| 2 | d2hxja1
|
|
 |
100.0 |
21 |
Fold:FinO-like
Superfamily:FinO-like
Family:FinO-like |
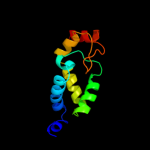
| 3 | c2hxjF_
|
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a protein of unknown function nmb1681 from2 neisseria meningitidis mc58, possible nucleic acid binding protein
|
| 4 | c2e70A_
|
|
 |
91.3 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt5;
PDBTitle: solution structure of the fifth kow motif of human2 transcription elongation factor spt5
|
| 5 | c3qiiA_
|
|
 |
88.1 |
20 |
PDB header:transcription regulator
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
|
| 6 | c3p8dB_
|
|
 |
82.0 |
21 |
PDB header:protein binding
Chain: B: PDB Molecule:medulloblastoma antigen mu-mb-50.72;
PDBTitle: crystal structure of the second tudor domain of human phf20 (homodimer2 form)
|
| 7 | c2in0A_
|
|
 |
74.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease pi-mtui;
PDBTitle: crystal structure of mtu reca intein splicing domain
|
| 8 | c2k8iA_
|
|
 |
72.4 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: solution structure of e.coli slyd
|
| 9 | c2equA_
|
|
 |
70.9 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
|
| 10 | c2qqsB_
|
|
 |
67.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:jmjc domain-containing histone demethylation
PDBTitle: jmjd2a tandem tudor domains in complex with a trimethylated2 histone h4-k20 peptide
|
| 11 | d1nxza1
|
|
 |
66.8 |
31 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 12 | c1zd7B_
|
|
 |
61.7 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: 1.7 angstrom crystal structure of post-splicing form of a dnae intein2 from synechocystis sp. pcc 6803
|
| 13 | c3dlmA_
|
|
 |
61.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:histone-lysine n-methyltransferase setdb1;
PDBTitle: crystal structure of tudor domain of human histone-lysine n-2 methyltransferase setdb1
|
| 14 | d2ix0a3
|
|
 |
60.8 |
32 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 15 | d1kk1a2
|
|
 |
59.6 |
19 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain |
| 16 | c2kfwA_
|
|
 |
58.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase
PDBTitle: solution structure of full-length slyd from e.coli
|
| 17 | c3d0fA_
|
|
 |
57.8 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:penicillin-binding 1 transmembrane protein mrca;
PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
|
| 18 | d1s0ua2
|
|
 |
56.4 |
20 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain |
| 19 | d2qn6a2
|
|
 |
55.8 |
21 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family:EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain |
| 20 | d3bzka4
|
|
 |
55.0 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 21 | d1sroa_ |
|
not modelled |
54.9 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 22 | c2xdpA_ |
|
not modelled |
51.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific demethylase 4c;
PDBTitle: crystal structure of the tudor domain of human jmjd2c
|
| 23 | d1at0a_ |
|
not modelled |
48.5 |
18 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Hedgehog C-terminal (Hog) autoprocessing domain |
| 24 | c2k52A_ |
|
not modelled |
47.3 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1198;
PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
|
| 25 | d1kl9a2 |
|
not modelled |
44.0 |
26 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 26 | d2do3a1 |
|
not modelled |
43.2 |
22 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:SPT5 KOW domain-like |
| 27 | d1ib8a1 |
|
not modelled |
40.9 |
24 |
Fold:Sm-like fold
Superfamily:YhbC-like, C-terminal domain
Family:YhbC-like, C-terminal domain |
| 28 | d1biaa2 |
|
not modelled |
38.6 |
15 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:Biotin repressor (BirA) |
| 29 | c2kvqG_ |
|
not modelled |
36.9 |
18 |
PDB header:transcription
Chain: G: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: solution structure of nuse:nusg-ctd complex
|
| 30 | c2jvvA_ |
|
not modelled |
36.9 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: solution structure of e. coli nusg carboxyterminal domain
|
| 31 | d1vqoq1 |
|
not modelled |
36.7 |
29 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
| 32 | d1kk8a1 |
|
not modelled |
35.7 |
13 |
Fold:SH3-like barrel
Superfamily:Myosin S1 fragment, N-terminal domain
Family:Myosin S1 fragment, N-terminal domain |
| 33 | d1whma_ |
|
not modelled |
33.5 |
26 |
Fold:SH3-like barrel
Superfamily:Cap-Gly domain
Family:Cap-Gly domain |
| 34 | c2khiA_ |
|
not modelled |
32.8 |
20 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
|
| 35 | c2keqA_ |
|
not modelled |
31.8 |
9 |
PDB header:splicing
Chain: A: PDB Molecule:dna polymerase iii alpha subunit, nucleic acid
PDBTitle: solution structure of dnae intein from nostoc punctiforme
|
| 36 | c2khjA_ |
|
not modelled |
31.8 |
24 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
|
| 37 | c3p8bB_ |
|
not modelled |
30.0 |
25 |
PDB header:transferase/transcription
Chain: B: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
|
| 38 | d1nppa2 |
|
not modelled |
29.5 |
21 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
| 39 | c2ix1A_ |
|
not modelled |
28.4 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:exoribonuclease 2;
PDBTitle: rnase ii d209n mutant
|
| 40 | c2hbpA_ |
|
not modelled |
28.3 |
24 |
PDB header:endocytosis, protein binding
Chain: A: PDB Molecule:cytoskeleton assembly control protein sla1;
PDBTitle: solution structure of sla1 homology domain 1
|
| 41 | c3q9qB_ |
|
not modelled |
26.6 |
19 |
PDB header:chaperone
Chain: B: PDB Molecule:heat shock protein beta-1;
PDBTitle: hspb1 fragment second crystal form
|
| 42 | d1wgka_ |
|
not modelled |
26.6 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:C9orf74 homolog |
| 43 | c1ssfA_ |
|
not modelled |
26.4 |
9 |
PDB header:cell cycle
Chain: A: PDB Molecule:transformation related protein 53 binding
PDBTitle: solution structure of the mouse 53bp1 fragment (residues2 1463-1617)
|
| 44 | c1zeqX_ |
|
not modelled |
25.9 |
17 |
PDB header:metal binding protein
Chain: X: PDB Molecule:cation efflux system protein cusf;
PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
|
| 45 | c2egwB_ |
|
not modelled |
25.5 |
9 |
PDB header:rna methyltransferase
Chain: B: PDB Molecule:upf0088 protein aq_165;
PDBTitle: crystal structure of rrna methyltransferase with sah ligand
|
| 46 | c2ckkA_ |
|
not modelled |
25.1 |
16 |
PDB header:nuclear protein
Chain: A: PDB Molecule:kin17;
PDBTitle: high resolution crystal structure of the human kin172 c-terminal domain containing a kow motif3 kin17.
|
| 47 | c2cghB_ |
|
not modelled |
24.9 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:biotin ligase;
PDBTitle: crystal structure of biotin ligase from mycobacterium2 tuberculosis
|
| 48 | c1vhkA_ |
|
not modelled |
24.9 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yqeu;
PDBTitle: crystal structure of an hypothetical protein
|
| 49 | c2imzA_ |
|
not modelled |
23.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease pi-mtui;
PDBTitle: crystal structure of mtu reca intein splicing domain
|
| 50 | d1nz9a_ |
|
not modelled |
23.4 |
29 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
| 51 | c2eayB_ |
|
not modelled |
23.0 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:biotin [acetyl-coa-carboxylase] ligase;
PDBTitle: crystal structure of biotin protein ligase from aquifex2 aeolicus
|
| 52 | d1mi8a_ |
|
not modelled |
23.0 |
16 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Intein (protein splicing domain) |
| 53 | c1kl9A_ |
|
not modelled |
21.3 |
26 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 2 subunit 1;
PDBTitle: crystal structure of the n-terminal segment of human eukaryotic2 initiation factor 2alpha
|
| 54 | d2ba0a1 |
|
not modelled |
20.9 |
13 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 55 | c3kw2A_ |
|
not modelled |
20.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable r-rna methyltransferase;
PDBTitle: crystal structure of probable rrna-methyltransferase from2 porphyromonas gingivalis
|
| 56 | c1ywuA_ |
|
not modelled |
20.3 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa4608;
PDBTitle: solution nmr structure of pseudomonas aeruginosa protein pa4608.2 northeast structural genomics target pat7
|
| 57 | d1ywua1 |
|
not modelled |
20.3 |
9 |
Fold:Split barrel-like
Superfamily:PilZ domain-like
Family:PilZ domain |
| 58 | d2rdea1 |
|
not modelled |
20.1 |
17 |
Fold:Split barrel-like
Superfamily:PilZ domain-like
Family:PilZ domain |
| 59 | d2z0sa1 |
|
not modelled |
20.0 |
10 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 60 | c2wj7D_ |
|
not modelled |
19.8 |
14 |
PDB header:chaperone
Chain: D: PDB Molecule:alpha-crystallin b chain;
PDBTitle: human alphab crystallin
|
| 61 | d2i5ua1 |
|
not modelled |
19.8 |
10 |
Fold:DnaD domain-like
Superfamily:DnaD domain-like
Family:DnaD domain |
| 62 | c2kr7A_ |
|
not modelled |
19.8 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase slyd;
PDBTitle: solution structure of helicobacter pylori slyd
|
| 63 | d1vhka1 |
|
not modelled |
19.6 |
13 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 64 | d1q46a2 |
|
not modelled |
18.9 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 65 | d1alla_ |
|
not modelled |
18.8 |
26 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 66 | d2je6i1 |
|
not modelled |
18.3 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 67 | c2d7gD_ |
|
not modelled |
18.1 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:primosomal protein n;
PDBTitle: crystal structure of the aa complex of the n-terminal2 domain of pria
|
| 68 | c2l1tA_ |
|
not modelled |
17.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of the n-terminal domain of np_954075.1
|
| 69 | c2ej9A_ |
|
not modelled |
17.6 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:putative biotin ligase;
PDBTitle: crystal structure of biotin protein ligase from2 methanococcus jannaschii
|
| 70 | c2ewnA_ |
|
not modelled |
17.5 |
14 |
PDB header:ligase, transcription
Chain: A: PDB Molecule:bira bifunctional protein;
PDBTitle: ecoli biotin repressor with co-repressor analog
|
| 71 | c2kq6A_ |
|
not modelled |
17.2 |
26 |
PDB header:transport protein
Chain: A: PDB Molecule:polycystin-2;
PDBTitle: the structure of the ef-hand domain of polycystin-2 suggests a2 mechanism for ca2+-dependent regulation of polycystin-2 channel3 activity
|
| 72 | d2nn6h1 |
|
not modelled |
16.2 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 73 | d1go3e1 |
|
not modelled |
16.1 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 74 | c1xniI_ |
|
not modelled |
15.8 |
13 |
PDB header:cell cycle
Chain: I: PDB Molecule:tumor suppressor p53-binding protein 1;
PDBTitle: tandem tudor domain of 53bp1
|
| 75 | c2zrrA_ |
|
not modelled |
15.3 |
25 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:mundticin ks immunity protein;
PDBTitle: crystal structure of an immunity protein that contributes2 to the self-protection of bacteriocin-producing3 enterococcus mundtii 15-1a
|
| 76 | d1ixda_ |
|
not modelled |
15.0 |
21 |
Fold:SH3-like barrel
Superfamily:Cap-Gly domain
Family:Cap-Gly domain |
| 77 | c1i84V_ |
|
not modelled |
14.9 |
23 |
PDB header:contractile protein
Chain: V: PDB Molecule:smooth muscle myosin heavy chain;
PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
|
| 78 | d1b33a_ |
|
not modelled |
14.8 |
22 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 79 | c2jkuA_ |
|
not modelled |
14.6 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:propionyl-coa carboxylase alpha chain,
PDBTitle: crystal structure of the n-terminal region of the biotin2 acceptor domain of human propionyl-coa carboxylase
|
| 80 | c3a8jF_ |
|
not modelled |
14.6 |
0 |
PDB header:transferase/transport protein
Chain: F: PDB Molecule:glycine cleavage system h protein;
PDBTitle: crystal structure of et-ehred complex
|
| 81 | c3mxuA_ |
|
not modelled |
14.5 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycine cleavage system h protein;
PDBTitle: crystal structure of glycine cleavage system protein h from bartonella2 henselae
|
| 82 | c3iftA_ |
|
not modelled |
14.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycine cleavage system h protein;
PDBTitle: crystal structure of glycine cleavage system protein h from2 mycobacterium tuberculosis, using x-rays from the compact light3 source.
|
| 83 | d1cpcb_ |
|
not modelled |
13.8 |
22 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 84 | c3cgnA_ |
|
not modelled |
13.6 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: crystal structure of thermophilic slyd
|
| 85 | d1br2a1 |
|
not modelled |
13.3 |
25 |
Fold:SH3-like barrel
Superfamily:Myosin S1 fragment, N-terminal domain
Family:Myosin S1 fragment, N-terminal domain |
| 86 | d2ghra1 |
|
not modelled |
13.2 |
42 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:HTS-like |
| 87 | c1v1hB_ |
|
not modelled |
13.0 |
19 |
PDB header:adenovirus
Chain: B: PDB Molecule:fibritin, fiber protein;
PDBTitle: adenovirus fibre shaft sequence n-terminally fused to the2 bacteriophage t4 fibritin foldon trimerisation motif with3 a short linker
|
| 88 | d1gmea_ |
|
not modelled |
12.8 |
32 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
| 89 | d1kn1a_ |
|
not modelled |
12.4 |
26 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 90 | c2qjlA_ |
|
not modelled |
12.2 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:ubiquitin-related modifier 1;
PDBTitle: crystal structure of urm1
|
| 91 | c2zkrq_ |
|
not modelled |
12.0 |
21 |
PDB header:ribosomal protein/rna
Chain: Q: PDB Molecule:rna expansion segment es31 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 92 | c3aabA_ |
|
not modelled |
11.9 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:putative uncharacterized protein st1653;
PDBTitle: small heat shock protein hsp14.0 with the mutations of i120f and i122f2 in the form i crystal
|
| 93 | c1br4E_ |
|
not modelled |
11.8 |
23 |
PDB header:muscle protein
Chain: E: PDB Molecule:myosin;
PDBTitle: smooth muscle myosin motor domain-essential light chain2 complex with mgadp.bef3 bound at the active site
|
| 94 | d1hpca_ |
|
not modelled |
11.7 |
15 |
Fold:Barrel-sandwich hybrid
Superfamily:Single hybrid motif
Family:Biotinyl/lipoyl-carrier proteins and domains |
| 95 | d1xo3a_ |
|
not modelled |
11.6 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:C9orf74 homolog |
| 96 | c1vdeA_ |
|
not modelled |
11.3 |
20 |
PDB header:endonuclease
Chain: A: PDB Molecule:pi-scei;
PDBTitle: pi-scei, a homing endonuclease with protein splicing2 activity
|
| 97 | c1vftA_ |
|
not modelled |
11.1 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of l-cycloserine-bound form of alanine2 racemase from d-cycloserine-producing streptomyces3 lavendulae
|
| 98 | d1dq3a1 |
|
not modelled |
11.0 |
11 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Intein (protein splicing domain) |
| 99 | d2h50a1 |
|
not modelled |
10.8 |
32 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |