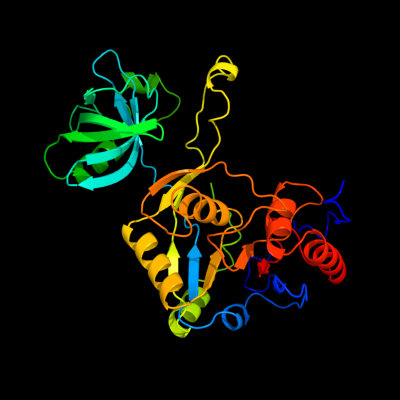
| 1 | c1yy3A_
|
|
 |
100.0 |
45 |
PDB header:isomerase
Chain: A: PDB Molecule:s-adenosylmethionine:trna ribosyltransferase-
PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
|
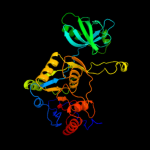
| 2 | d1wdia_
|
|
 |
100.0 |
48 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
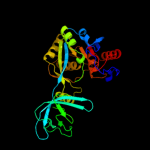
| 3 | d1vkya_
|
|
 |
100.0 |
46 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
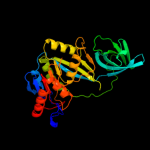
| 4 | d1jr7a_
|
|
 |
88.5 |
20 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Gab protein (hypothetical protein YgaT) |
| 5 | c3ms5A_
|
|
 |
86.1 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gamma-butyrobetaine dioxygenase;
PDBTitle: crystal structure of human gamma-butyrobetaine,2-oxoglutarate2 dioxygenase 1 (bbox1)
|
| 6 | d1ds1a_
|
|
 |
84.7 |
56 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Clavaminate synthase |
| 7 | c2og5A_
|
|
 |
83.4 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxygenase;
PDBTitle: crystal structure of asparagine oxygenase (asno)
|
| 8 | c2wbqA_
|
|
 |
81.9 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arginine beta-hydroxylase;
PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
|
| 9 | d1oiha_
|
|
 |
81.2 |
17 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 10 | d1otja_
|
|
 |
79.3 |
17 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 11 | c3r1jB_
|
|
 |
77.2 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
|
| 12 | c3pvjB_
|
|
 |
76.1 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of the fe(ii)/alpha-ketoglutarate dependent taurine2 dioxygenase from pseudomonas putida kt2440
|
| 13 | c3eatX_
|
|
 |
74.8 |
22 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:pyoverdine biosynthesis protein pvcb;
PDBTitle: crystal structure of the pvcb (pa2255) protein from2 pseudomonas aeruginosa
|
| 14 | d1y0za_
|
|
 |
69.3 |
33 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:gamma-Butyrobetaine hydroxylase |
| 15 | d1qyia_
|
|
 |
63.7 |
26 |
Fold:HAD-like
Superfamily:HAD-like
Family:Hypothetical protein MW1667 (SA1546) |
| 16 | d1nx4a_
|
|
 |
61.9 |
50 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:gamma-Butyrobetaine hydroxylase |
| 17 | c3p9zA_
|
|
 |
47.6 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 18 | c2nuxB_
|
|
 |
47.4 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
| 19 | d1hg3a_
|
|
 |
44.7 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 20 | c2hc9A_
|
|
 |
44.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:leucine aminopeptidase 1;
PDBTitle: structure of caenorhabditis elegans leucine aminopeptidase-zinc2 complex (lap1)
|
| 21 | d1a3xa1 |
|
not modelled |
44.5 |
11 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 22 | c3ny0D_ |
|
not modelled |
43.2 |
14 |
PDB header:metal binding protein
Chain: D: PDB Molecule:urease accessory protein uree;
PDBTitle: crystal structure of uree from helicobacter pylori (ni2+ bound form)
|
| 23 | c3pihA_ |
|
not modelled |
42.8 |
32 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:uvrabc system protein a;
PDBTitle: t. maritima uvra in complex with fluorescein-modified dna
|
| 24 | d1ttja_ |
|
not modelled |
40.1 |
45 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 25 | c2h6rG_ |
|
not modelled |
40.0 |
19 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
| 26 | d1jr2a_ |
|
not modelled |
38.6 |
20 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 27 | c1jr2A_ |
|
not modelled |
38.6 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: structure of uroporphyrinogen iii synthase
|
| 28 | d1n55a_ |
|
not modelled |
37.4 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 29 | c2vf7B_ |
|
not modelled |
37.2 |
39 |
PDB header:dna-binding protein
Chain: B: PDB Molecule:excinuclease abc, subunit a.;
PDBTitle: crystal structure of uvra2 from deinococcus radiodurans
|
| 30 | d1mo0a_ |
|
not modelled |
37.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 31 | d1pkla1 |
|
not modelled |
35.5 |
15 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 32 | d1w0ma_ |
|
not modelled |
35.5 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 33 | d2ebfx2 |
|
not modelled |
34.9 |
10 |
Fold:EreA/ChaN-like
Superfamily:EreA/ChaN-like
Family:PMT domain-like |
| 34 | d1wjja_ |
|
not modelled |
34.6 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 35 | d2evra2 |
|
not modelled |
34.5 |
28 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 36 | d1sw3a_ |
|
not modelled |
33.4 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 37 | c2dp3A_ |
|
not modelled |
32.9 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
|
| 38 | c1pklB_ |
|
not modelled |
32.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyruvate kinase);
PDBTitle: the structure of leishmania pyruvate kinase
|
| 39 | d1wd7a_ |
|
not modelled |
31.0 |
18 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 40 | d2b8ea1 |
|
not modelled |
30.1 |
17 |
Fold:HAD-like
Superfamily:HAD-like
Family:Meta-cation ATPase, catalytic domain P |
| 41 | c3fkkA_ |
|
not modelled |
29.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
| 42 | c1yyaA_ |
|
not modelled |
29.4 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
|
| 43 | d1omha_ |
|
not modelled |
28.7 |
31 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:Relaxase domain |
| 44 | d1neya_ |
|
not modelled |
28.6 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 45 | c3kxqB_ |
|
not modelled |
28.4 |
45 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from bartonella2 henselae at 1.6a resolution
|
| 46 | c3i5aA_ |
|
not modelled |
28.2 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator/ggdef domain protein;
PDBTitle: crystal structure of full-length wpsr from pseudomonas syringae
|
| 47 | c3hvbB_ |
|
not modelled |
28.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein fimx;
PDBTitle: crystal structure of the dual-domain ggdef-eal module of2 fimx from pseudomonas aeruginosa
|
| 48 | c3l6tB_ |
|
not modelled |
28.2 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:mobilization protein trai;
PDBTitle: crystal structure of an n-terminal mutant of the plasmid pcu1 trai2 relaxase domain
|
| 49 | d1b9ba_ |
|
not modelled |
28.1 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 50 | c3d8tB_ |
|
not modelled |
28.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
|
| 51 | d1diha1 |
|
not modelled |
27.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 52 | c2x7jA_ |
|
not modelled |
26.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: structure of the menaquinone biosynthesis protein mend from2 bacillus subtilis
|
| 53 | d1r2ra_ |
|
not modelled |
26.7 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 54 | d1suxa_ |
|
not modelled |
26.6 |
42 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 55 | d1o7ia_ |
|
not modelled |
26.2 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 56 | d1kv5a_ |
|
not modelled |
25.8 |
45 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 57 | c1nl3B_ |
|
not modelled |
25.8 |
31 |
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase seca 1 subunit;
PDBTitle: crystal structure of the seca protein translocation atpase2 from mycobacterium tuberculosis in apo form
|
| 58 | d1xnea_ |
|
not modelled |
24.4 |
33 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 59 | d1w3ia_ |
|
not modelled |
23.7 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 60 | c1ayeA_ |
|
not modelled |
23.4 |
16 |
PDB header:serine protease
Chain: A: PDB Molecule:procarboxypeptidase a2;
PDBTitle: human procarboxypeptidase a2
|
| 61 | c3m9yB_ |
|
not modelled |
22.5 |
35 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from methicillin2 resistant staphylococcus aureus at 1.9 angstrom resolution
|
| 62 | c3civA_ |
|
not modelled |
22.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-mannanase;
PDBTitle: crystal structure of the endo-beta-1,4-mannanase from2 alicyclobacillus acidocaldarius
|
| 63 | c2gm2A_ |
|
not modelled |
21.9 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
|
| 64 | c3ijpA_ |
|
not modelled |
21.7 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase from2 bartonella henselae at 2.0a resolution
|
| 65 | c2jgqB_ |
|
not modelled |
21.6 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: kinetics and structural properties of triosephosphate2 isomerase from helicobacter pylori
|
| 66 | c3shoA_ |
|
not modelled |
20.7 |
29 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, rpir family;
PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
|
| 67 | c3dinB_ |
|
not modelled |
20.5 |
25 |
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:protein translocase subunit seca;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
|
| 68 | d1jmca1 |
|
not modelled |
20.5 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 69 | d1m6ja_ |
|
not modelled |
20.5 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 70 | c2boaB_ |
|
not modelled |
20.5 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:carboxypeptidase a4;
PDBTitle: human procarboxypeptidase a4.
|
| 71 | d2g50a1 |
|
not modelled |
20.4 |
8 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 72 | c2k50A_ |
|
not modelled |
20.0 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:replication factor a related protein;
PDBTitle: solution nmr structure of the replication factor a related2 protein from methanobacterium thermoautotrophicum.3 northeast structural genomics target tr91a.
|
| 73 | d1ntga_ |
|
not modelled |
19.9 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Myf domain |
| 74 | c3bi8A_ |
|
not modelled |
19.9 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 75 | d1o5xa_ |
|
not modelled |
19.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 76 | c1drwA_ |
|
not modelled |
19.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
| 77 | d1hl2a_ |
|
not modelled |
19.2 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 78 | c2eo2A_ |
|
not modelled |
18.4 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:adult male hypothalamus cdna, riken full-length
PDBTitle: solution structure of the insertion region (510-573) of2 fthfs domain from mouse methylenetetrahydrofolate3 dehydrogenase (nadp+ dependent) 1-like protein
|
| 79 | c3s6dA_ |
|
not modelled |
18.3 |
32 |
PDB header:isomerase
Chain: A: PDB Molecule:putative triosephosphate isomerase;
PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
|
| 80 | c3lerA_ |
|
not modelled |
18.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 81 | c1ynxA_ |
|
not modelled |
18.0 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:replication factor-a protein 1;
PDBTitle: solution structure of dna binding domain a (dbd-a) of2 s.cerevisiae replication protein a (rpa)
|
| 82 | c3krsB_ |
|
not modelled |
17.9 |
32 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
| 83 | c3gvgA_ |
|
not modelled |
17.6 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from mycobacterium2 tuberculosis
|
| 84 | c2zpmA_ |
|
not modelled |
16.9 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
|
| 85 | d1o1xa_ |
|
not modelled |
16.8 |
25 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
| 86 | c3th6B_ |
|
not modelled |
16.8 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|
| 87 | c2ygrD_ |
|
not modelled |
16.7 |
36 |
PDB header:hydrolase
Chain: D: PDB Molecule:uvrabc system protein a;
PDBTitle: mycobacterium tuberculosis uvra
|
| 88 | c3pl0B_ |
|
not modelled |
16.2 |
25 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a bsma homolog (mpe_a2762) from methylobium2 petroleophilum pm1 at 1.91 a resolution
|
| 89 | d1trea_ |
|
not modelled |
16.2 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 90 | c3ahxC_ |
|
not modelled |
16.1 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase a;
PDBTitle: crystal structure of beta-glucosidase a from bacterium clostridium2 cellulovorans
|
| 91 | d1p4da_ |
|
not modelled |
16.0 |
28 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:Relaxase domain |
| 92 | d1aw1a_ |
|
not modelled |
15.8 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 93 | d1wcga1 |
|
not modelled |
15.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 94 | d1owfa_ |
|
not modelled |
15.6 |
22 |
Fold:IHF-like DNA-binding proteins
Superfamily:IHF-like DNA-binding proteins
Family:Prokaryotic DNA-bending protein |
| 95 | c3hv9A_ |
|
not modelled |
15.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein fimx;
PDBTitle: crystal structure of fimx eal domain from pseudomonas aeruginosa
|
| 96 | c3e96B_ |
|
not modelled |
15.4 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 97 | d1s04a_ |
|
not modelled |
15.4 |
27 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 98 | c2iyeC_ |
|
not modelled |
15.3 |
23 |
PDB header:hydrolase
Chain: C: PDB Molecule:copper-transporting atpase;
PDBTitle: structure of catalytic cpx-atpase domain copb-b
|
| 99 | d1o5ka_ |
|
not modelled |
14.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |