1 c3r1rB_
100.0
100
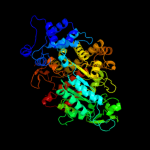
PDB header: complex (oxidoreductase/peptide)Chain: B: PDB Molecule: ribonucleotide reductase r1 protein;PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
2 c3hnfA_
100.0
30
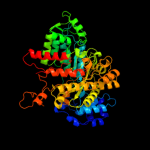
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase large subunit;PDBTitle: crystal structure of human ribonucleotide reductase 1 bound to the2 effectors ttp and datp
3 c2wghA_
100.0
30
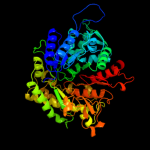
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase largePDBTitle: human ribonucleotide reductase r1 subunit (rrm1) in complex2 with datp and mg.
4 c1pemA_
100.0
22
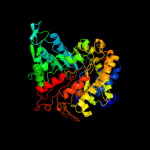
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase 2 alphaPDBTitle: ribonucleotide reductase protein r1e from salmonella2 typhimurium
5 c2cvuA_
100.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase large chainPDBTitle: structures of yeast ribonucleotide reductase i
6 d1rlra2
100.0
99
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: R1 subunit of ribonucleotide reductase, C-terminal domain7 c1xjeA_
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleotide reductase, b12-dependent;PDBTitle: structural mechanism of allosteric substrate specificity in a2 ribonucleotide reductase: dttp-gdp complex
8 d1peqa2
100.0
25
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: R1 subunit of ribonucleotide reductase, C-terminal domain9 d1l1la_
100.0
15
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: B12-dependent (class II) ribonucleotide reductase10 d1rlra1
100.0
100
Fold: R1 subunit of ribonucleotide reductase, N-terminal domainSuperfamily: R1 subunit of ribonucleotide reductase, N-terminal domainFamily: R1 subunit of ribonucleotide reductase, N-terminal domain11 d1peqa1
100.0
14
Fold: R1 subunit of ribonucleotide reductase, N-terminal domainSuperfamily: R1 subunit of ribonucleotide reductase, N-terminal domainFamily: R1 subunit of ribonucleotide reductase, N-terminal domain12 d1hk8a_
95.3
13
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit13 c1hk8A_
95.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
14 d1h16a_
86.4
19
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like15 d1qhma_
43.8
17
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like16 c2f3oB_
43.3
19
PDB header: unknown functionChain: B: PDB Molecule: pyruvate formate-lyase 2;PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
17 d1r9da_
42.8
16
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like18 d1h3ob_
34.1
13
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs19 d2okqa1
33.0
8
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: YbaA-like20 d1vjfa_
31.5
18
Fold: YbaK/ProRS associated domainSuperfamily: YbaK/ProRS associated domainFamily: YbaK/ProRS associated domain21 c2okqB_
not modelled
22.8
8
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ybaa;PDBTitle: crystal structure of unknown conserved ybaa protein from2 shigella flexneri
22 c2bjgB_
not modelled
22.6
12
PDB header: hydrolaseChain: B: PDB Molecule: choloylglycine hydrolase;PDBTitle: crystal structure of conjugated bile acid hydrolase from2 clostridium perfringens in complex with reaction products3 taurine and deoxycholate
23 c2hezB_
not modelled
20.8
14
PDB header: hydrolaseChain: B: PDB Molecule: bile salt hydrolase;PDBTitle: bifidobacterium longum bile salt hydrolase
24 d2pvaa_
not modelled
19.7
12
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Penicillin V acylase25 d1j09a2
not modelled
19.2
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain26 d2ccqa1
not modelled
18.5
16
Fold: PUG domain-likeSuperfamily: PUG domain-likeFamily: PUG domain27 c1pgjA_
not modelled
14.7
39
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase;PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
28 c3pg6D_
not modelled
14.5
15
PDB header: ligaseChain: D: PDB Molecule: e3 ubiquitin-protein ligase dtx3l;PDBTitle: the carboxyl terminal domain of human deltex 3-like
29 d2b4va2
not modelled
13.7
44
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: RNA editing terminal uridyl transferase 2, RET2, catalytic domain30 c2iz1C_
not modelled
13.7
28
PDB header: oxidoreductaseChain: C: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating;PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
31 d1vmha_
not modelled
12.1
13
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like32 d1vmfa_
not modelled
12.1
16
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like33 d1ywsa1
not modelled
12.1
21
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger34 d1mska_
not modelled
12.1
20
Fold: Methionine synthase activation domain-likeSuperfamily: Methionine synthase activation domain-likeFamily: Methionine synthase SAM-binding domain35 c2p4qA_
not modelled
12.0
33
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating 1;PDBTitle: crystal structure analysis of gnd1 in saccharomyces cerevisiae
36 c2jr7A_
not modelled
11.6
21
PDB header: metal binding proteinChain: A: PDB Molecule: dph3 homolog;PDBTitle: solution structure of human desr1
37 d1vmja_
not modelled
11.5
26
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like38 d1wgea1
not modelled
11.2
21
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger39 d1avca2
not modelled
10.9
14
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin40 d1vkia_
not modelled
10.5
13
Fold: YbaK/ProRS associated domainSuperfamily: YbaK/ProRS associated domainFamily: YbaK/ProRS associated domain41 c2p6hB_
not modelled
10.3
26
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of hypothetical protein ape1520 from aeropyrum2 pernix k1
42 c2xrgA_
not modelled
10.3
11
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
43 c2oqcB_
not modelled
10.3
10
PDB header: hydrolaseChain: B: PDB Molecule: penicillin v acylase;PDBTitle: crystal structure of penicillin v acylase from bacillus subtilis
44 d1xn7a_
not modelled
10.3
64
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Hypothetical protein YhgG45 d2d0ob1
not modelled
9.6
10
Fold: Anticodon-binding domain-likeSuperfamily: B12-dependent dehydatase associated subunitFamily: Dehydratase-reactivating factor beta subunit46 c3rcqA_
not modelled
9.1
25
PDB header: oxidoreductaseChain: A: PDB Molecule: aspartyl/asparaginyl beta-hydroxylase;PDBTitle: crystal structure of human aspartate beta-hydroxylase isoform a
47 c2qlwA_
not modelled
8.9
20
PDB header: isomeraseChain: A: PDB Molecule: rhau;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
48 c2qlxA_
not modelled
8.9
20
PDB header: isomeraseChain: A: PDB Molecule: l-rhamnose mutarotase;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
49 d1vpha_
not modelled
8.7
14
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like50 c2p6cB_
not modelled
8.5
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: aq_2013 protein;PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
51 d1wiga2
not modelled
8.3
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain52 d2g5ca2
not modelled
8.3
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain53 c3dzbA_
not modelled
8.2
26
PDB header: biosynthetic proteinChain: A: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
54 c2xr9A_
not modelled
7.8
11
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2)
55 d1f6da_
not modelled
7.7
18
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase56 d1x8da1
not modelled
7.5
40
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: YiiL-like57 c1vraA_
not modelled
7.4
10
PDB header: transferaseChain: A: PDB Molecule: arginine biosynthesis bifunctional protein argj;PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
58 c2g5cD_
not modelled
7.4
32
PDB header: oxidoreductaseChain: D: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
59 d1gv2a2
not modelled
7.4
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain60 d1f0ya2
not modelled
7.3
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain61 c2pv7B_
not modelled
7.2
39
PDB header: isomerase, oxidoreductaseChain: B: PDB Molecule: t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
62 c3sggA_
not modelled
7.1
7
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical hydrolase;PDBTitle: crystal structure of a hypothetical hydrolase (bt_2193) from2 bacteroides thetaiotaomicron vpi-5482 at 1.25 a resolution
63 c1idzA_
not modelled
7.0
24
PDB header: dna-binding proteinChain: A: PDB Molecule: mouse c-myb dna-binding domain repeat 3;PDBTitle: structure of myb transforming protein, nmr, 20 structures
64 c3kowH_
not modelled
6.9
20
PDB header: metal binding proteinChain: H: PDB Molecule: d-ornithine aminomutase s component;PDBTitle: crystal structure of ornithine 4,5 aminomutase backsoaked complex
65 c3ggpA_
not modelled
6.8
32
PDB header: oxidoreductaseChain: A: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
66 d1dm5a_
not modelled
6.8
19
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin67 d1at3a_
not modelled
6.7
30
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin68 d2csba2
not modelled
6.7
21
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain69 c2wasA_
not modelled
6.6
25
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase;PDBTitle: structure of the fungal type i fas ppt domain
70 d1wdka3
not modelled
6.6
39
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain71 d2pgda2
not modelled
6.6
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain72 d1hm6a_
not modelled
6.5
18
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin73 d1nbwb_
not modelled
6.4
25
Fold: Anticodon-binding domain-likeSuperfamily: B12-dependent dehydatase associated subunitFamily: Dehydratase-reactivating factor beta subunit74 c2ep9A_
not modelled
6.3
33
PDB header: oxidoreductaseChain: A: PDB Molecule: l-gulonate 3-dehydrogenase;PDBTitle: crystal structure of the rabbit l-gulonate 3-dehydrogenase2 (nadh form)
75 d1chua3
not modelled
6.2
15
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain76 c3crw1_
not modelled
6.2
16
PDB header: hydrolaseChain: 1: PDB Molecule: xpd/rad3 related dna helicase;PDBTitle: "xpd_apo"
77 d1eexb_
not modelled
6.1
22
Fold: Anticodon-binding domain-likeSuperfamily: B12-dependent dehydatase associated subunitFamily: Diol dehydratase, beta subunit78 d1vzva_
not modelled
5.9
30
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin79 c2c1fA_
not modelled
5.9
22
PDB header: hydrolaseChain: A: PDB Molecule: bifunctional endo-1,4-beta-xylanase a;PDBTitle: the structure of the family 11 xylanase from neocallimastix2 patriciarum
80 d1i36a2
not modelled
5.9
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain81 c2h1zA_
not modelled
5.9
50
PDB header: toxinChain: A: PDB Molecule: hybrid atracotoxin;PDBTitle: structure of a dual-target spider toxin
82 d1wo8a1
not modelled
5.8
13
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA83 d1hrti_
not modelled
5.8
40
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Hirudin-like84 c2v4iA_
not modelled
5.8
6
PDB header: transferaseChain: A: PDB Molecule: glutamate n-acetyltransferase 2 alpha chain;PDBTitle: structure of a novel n-acyl-enzyme intermediate of an n-2 terminal nucleophile (ntn) hydrolase, oat2
85 d1zeta1
not modelled
5.8
20
Fold: Lesion bypass DNA polymerase (Y-family), little finger domainSuperfamily: Lesion bypass DNA polymerase (Y-family), little finger domainFamily: Lesion bypass DNA polymerase (Y-family), little finger domain86 d3cuma2
not modelled
5.8
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain87 c2dwuA_
not modelled
5.8
15
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
88 d1mbja_
not modelled
5.8
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain89 c1mbjA_
not modelled
5.8
21
PDB header: dna binding proteinChain: A: PDB Molecule: myb proto-oncogene protein;PDBTitle: mouse c-myb dna-binding domain repeat 3
90 c2rpaA_
not modelled
5.8
12
PDB header: hydrolaseChain: A: PDB Molecule: katanin p60 atpase-containing subunit a1;PDBTitle: the solution structure of n-terminal domain of microtubule severing2 enzyme
91 d1sfkb_
not modelled
5.7
20
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C92 d1uoua1
not modelled
5.7
18
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain93 c3njqB_
not modelled
5.7
30
PDB header: viral protein/inhibitorChain: B: PDB Molecule: orf 17;PDBTitle: crystal structure of kaposi's sarcoma-associated herpesvirus protease2 in complex with dimer disruptor
94 d1m9ia2
not modelled
5.7
22
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin95 c2p6uA_
not modelled
5.7
36
PDB header: dna binding proteinChain: A: PDB Molecule: afuhel308 helicase;PDBTitle: apo structure of the hel308 superfamily 2 helicase
96 c2rafC_
not modelled
5.7
17
PDB header: oxidoreductaseChain: C: PDB Molecule: putative dinucleotide-binding oxidoreductase;PDBTitle: crystal structure of putative dinucleotide-binding2 oxidoreductase (np_786167.1) from lactobacillus plantarum3 at 1.60 a resolution
97 d1h4ga_
not modelled
5.6
17
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Xylanase/endoglucanase 11/1298 d2pv7a2
not modelled
5.6
44
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain99 c2ysrA_
not modelled
5.6
17
PDB header: signaling proteinChain: A: PDB Molecule: dep domain-containing protein 1;PDBTitle: solution structure of the dep domain from human dep domain-2 containing protein 1